Abstract
Purpose
Idelalisib is a potent PI3Kδ inhibitor that was recently approved for treating hematologic malignancies. The objective of this analysis was to develop a population pharmacokinetic model for idelalisib and its inactive metabolite GS-563117 and to evaluate the impact of covariates on idelalisib/GS-563117 PK.
Methods
Data from 10 phase I or II studies in healthy volunteers or patients with hematologic malignancies (n = 736) were analyzed using NONMEM. Stepwise forward addition followed by backward elimination was implemented in the covariate (age, gender, race, body weight, baseline CLcr, AST, ALT, disease status, and type of cancer) model building process. Various model assessment methods were used to evaluate the models.
Results
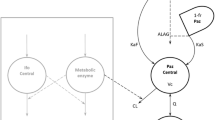
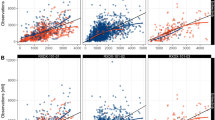
Idelalisib plasma PK was best described by a two-compartment model with first-order absorption, first-order elimination from the central compartment, and a lag time. A nonlinear relationship between dose and relative bioavailability was included in the final model. Two statistically significant covariates were identified and incorporated into the final model: health status (healthy vs. patient) on CL/F and Q/F and body weight on CL/F. Despite being a statistically significant covariate, the effect of body weight on idelalisib exposures was weak, as evidenced by minor changes of steady-state exposure (C trough: 16 %; AUC and C max: 10 %) for a patient with extreme body weight (5th and 95th percentile) relative to the typical patient, and not considered to be clinically relevant.
Conclusions
PopPK models were developed to adequately describe the plasma concentrations of idelalisib and GS-563117. There were no covariate that had a clinically meaningful impact on idelalisib or GS-563117 exposure.



Similar content being viewed by others
References
Okkenhaug K, Vanhaesebroeck B (2003) PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol 3(4):317–330. doi:10.1038/nri1056
Vanhaesebroeck B, Ali K, Bilancio A, Geering B, Foukas LC (2005) Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem Sci 30(4):194–204. doi:10.1016/j.tibs.2005.02.008
Herman SE, Lapalombella R, Gordon AL, Ramanunni A, Blum KA, Jones J, Zhang X, Lannutti BJ, Puri KD, Muthusamy N, Byrd JC, Johnson AJ (2011) The role of phosphatidylinositol 3-kinase-delta in the immunomodulatory effects of lenalidomide in chronic lymphocytic leukemia. Blood 117(16):4323–4327. doi:10.1182/blood-2010-11-315705
Bernal A, Pastore RD, Asgary Z, Keller SA, Cesarman E, Liou HC, Schattner EJ (2001) Survival of leukemic B cells promoted by engagement of the antigen receptor. Blood 98(10):3050–3057
Lannutti BJ, Meadows SA, Herman SE, Kashishian A, Steiner B, Johnson AJ, Byrd JC, Tyner JW, Loriaux MM, Deininger M, Druker BJ, Puri KD, Ulrich RG, Giese NA (2011) CAL-101, a p110delta selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood 117(2):591–594. doi:10.1182/blood-2010-03-275305
Hoellenriegel J, Meadows SA, Sivina M, Wierda WG, Kantarjian H, Keating MJ, Giese N, O’Brien S, Yu A, Miller LL, Lannutti BJ, Burger JA (2011) The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood 118(13):3603–3612. doi:10.1182/blood-2011-05-352492
Gopal AK, Kahl BS, de Vos S, Wagner-Johnston ND, Schuster SJ, Jurczak WJ, Flinn IW, Flowers CR, Martin P, Viardot A, Blum KA, Goy AH, Davies AJ, Zinzani PL, Dreyling M, Johnson D, Miller LL, Holes L, Li D, Dansey RD, Godfrey WR, Salles GA (2014) PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med 370(11):1008–1018. doi:10.1056/NEJMoa1314583
Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn I, Ghia P, Eradat H, Ervin T, Lamanna N, Coiffier B, Pettitt AR, Ma S, Stilgenbauer S, Cramer P, Aiello M, Johnson DM, Miller LL, Li D, Jahn TM, Dansey RD, Hallek M, O’Brien SM (2014) Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med 370(11):997–1007. doi:10.1056/NEJMoa1315226
Beals SL, Boeckmann AJ, Sheiner LB (1988–1992) NONMEM user’s guide, part I–VII
Lindbom L, Pihlgren P, Jonsson EN (2005) PsN-Toolkit—a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Progr Biomed 79(3):241–257. doi:10.1016/j.cmpb.2005.04.005
Lindbom L, Ribbing J, Jonsson EN (2004) Perl-speaks-NONMEM (PsN)—a perl module for NONMEM related programming. Comput Methods Progr Biomed 75(2):85–94. doi:10.1016/j.cmpb.2003.11.003
Karlsson MO, Holford, N. A tutorial on visual predictive checks. In: PAGE 17, 2008. Abstr 1434 www.page-meeting.org/?abstract=1434
Harling K, Uekcert SA (2011) NPC/VPC userguide and technical description. 2011 05-26 PsN 3.4.2
Ette EI (1997) Stability and performance of a population pharmacokinetic model. J Clin Pharmacol 37(6):486–495
Jin F, Zhou H, Holes L, Li X, Newcomb T, Dansey R, Ramananthan S (2014) Exposure-response of idelalisib, a novel Pi3kδ inhibitor, in the treatment of hematologic malignancies. American Society of Clinical Pharmacology and Therapeutics, Atlanta, GA, LBI-002
Jin F, Robeson M, Zhou H, Hisoire G, Ramanathan S (2014) The pharmacokinetics and safety of idelalisib in subjects with severe renal impairment. American Association for Cancer Research, San Diego, CA, 2014. CT204
Jin F, Robeson M, Zhou H, Kwan E, Ramananthan S (2014) Pharmacokinetics, metabolism and excretion of idelalisib. American Conference of Cancer Research, San Diego, CA, 2014. Abstract 4633
Jin F, Robeson M, Zhou H, Hisoire G, Ramanathan S (2014) The pharmacokinetics and safety of idelalisib in subjects with moderate or severe hepatic impairment. American Society of Clinical Oncology. IL, Chicago, p 2592
Acknowledgments
The results of this study were presented in part at the 2014 American Conference of Pharmacometric (ACOP) (October 13, 2014, Las Vegas, NV). Financial support for this study was provided by Gilead Sciences, Inc.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jin, F., Gao, Y., Zhou, H. et al. Population pharmacokinetic modeling of idelalisib, a novel PI3Kδ inhibitor, in healthy subjects and patients with hematologic malignancies. Cancer Chemother Pharmacol 77, 89–98 (2016). https://doi.org/10.1007/s00280-015-2891-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-015-2891-8