Abstract
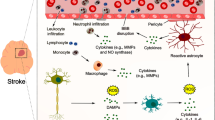
Ischemic stroke (IS) is one of the most impairing complications of sickle cell anemia (SCA), responsible for 20% of mortality in patients. Rheological alterations, adhesive properties of sickle reticulocytes, leukocyte adhesion, inflammation and endothelial dysfunction are related to the vasculopathy observed prior to ischemic events. The role of the vascular endothelium in this complex cascade of mechanisms is emphasized, as well as in the process of ischemia-induced repair and neovascularization. The aim of the present study was to perform a comparative transcriptomic analysis of endothelial colony-forming cells (ECFCs) from SCA patients with and without IS. Next, to gain further insights of the biological relevance of differentially expressed genes (DEGs), functional enrichment analysis, protein–protein interaction network (PPI) construction and in silico prediction of regulatory factors were performed. Among the 2469 DEGs, genes related to cell proliferation (AKT1, E2F1, CDCA5, EGFL7), migration (AKT1, HRAS), angiogenesis (AKT1, EGFL7) and defense response pathways (HRAS, IRF3, TGFB1), important endothelial cell molecular mechanisms in post ischemia repair were identified. Despite the severity of IS in SCA, widely accepted molecular targets are still lacking, especially related to stroke outcome. The comparative analysis of the gene expression profile of ECFCs from IS patients versus controls seems to indicate that there is a persistent angiogenic process even after a long time this complication has occurred. Thus, this is an original study which may lead to new insights into the molecular basis of SCA stroke and contribute to a better understanding of the role of endothelial cells in stroke recovery.







Similar content being viewed by others
References
Kato GJ, Piel FB, Reid CD et al (2018) Sickle cell disease. Nat Rev Dis Primers 4:18010. https://doi.org/10.1038/nrdp.2018.10
Balkaran B, Char G, Morris JS et al (1992) Stroke in a cohort of patients with homozygous sickle cell disease. J Pediatr 120:360–366. https://doi.org/10.1016/S0022-3476(05)80897-2
Ohene-Frempong K, Weiner SJ, Sleeper LA et al (1998) Cerebrovascular accidents in sickle cell disease: rates and risk factors. Blood 91:288–294
Driscoll MC (2003) Stroke risk in siblings with sickle cell anemia. Blood 101:2401–2404. https://doi.org/10.1182/blood.V101.6.2401
Adams R, McKie V, Nichols F et al (1992) The use of transcranial ultrasonography to predict stroke in sickle cell disease. N Engl J Med 326:605–610. https://doi.org/10.1056/NEJM199202273260905
Adams RJ (2013) Toward a stroke-free childhood in sickle cell disease. Stroke 44:2930–2934. https://doi.org/10.1161/STROKEAHA.113.001312
Adams RJ, McKie VC, Hsu L et al (1998) Prevention of a first stroke by transfusions in children with sickle cell anemia and abnormal results on transcranial doppler ultrasonography. N Engl J Med 339:5–11. https://doi.org/10.1056/NEJM199807023390102
Ware RE, Davis BR, Schultz WH et al (2016) Hydroxycarbamide versus chronic transfusion for maintenance of transcranial doppler flow velocities in children with sickle cell anaemia—TCD with transfusions changing to hydroxyurea (TWiTCH): a multicentre, open-label, phase 3, non-inferiority trial. The Lancet 387:661–670. https://doi.org/10.1016/S0140-6736(15)01041-7
Runge A, Brazel D, Pakbaz Z (2022) Stroke in sickle cell disease and the promise of recent disease modifying agents. J Neurol Sci 442:120412. https://doi.org/10.1016/j.jns.2022.120412
Liao S, Luo C, Cao B et al (2017) Endothelial progenitor cells for ischemic stroke: update on basic research and application. Stem Cells Int 2017:1–12. https://doi.org/10.1155/2017/2193432
Moubarik C, Guillet B, Youssef B et al (2011) Transplanted late outgrowth endothelial progenitor cells as cell therapy product for stroke. Stem Cell Rev Rep 7:208–220. https://doi.org/10.1007/s12015-010-9157-y
Rakkar K, Othman O, Sprigg N et al (2020) Endothelial progenitor cells, potential biomarkers for diagnosis and prognosis of ischemic stroke: protocol for an observational case-control study. Neural Regen Res 15:1300. https://doi.org/10.4103/1673-5374.269028
Yan F, Liu X, Ding H, Zhang W (2022) Paracrine mechanisms of endothelial progenitor cells in vascular repair. Acta Histochem 124:151833. https://doi.org/10.1016/j.acthis.2021.151833
Shirota T, He H, Yasui H, Matsuda T (2003) Human endothelial progenitor cell-seeded hybrid graft: proliferative and antithrombogenic potentials in vitro and fabrication processing. Tissue Eng 9:127–136. https://doi.org/10.1089/107632703762687609
Alwjwaj M, Kadir RA, Bayraktutan U (2021) The secretome of endothelial progenitor cells: a potential therapeutic strategy for ischemic stroke. Neural Regen Res 16:1483. https://doi.org/10.4103/1673-5374.303012
Hur J, Yoon C-H, Kim H-S et al (2004) Characterization of two types of endothelial progenitor cells and their different contributions to neovasculogenesis. Arterioscler Thromb Vasc Biol 24:288–293. https://doi.org/10.1161/01.ATV.0000114236.77009.06
Ito MT, da Silva Costa SM, Baptista LC, et al (2020) Angiogenesis-related genes in endothelial progenitor cells may be involved in sickle cell stroke. J Am Heart Assoc 9(3):e014143. https://doi.org/10.1161/JAHA.119.014143
de Bertozzo VHE, da Silva Costa SM, Ito MT et al (2023) Comparative transcriptome analysis of endothelial progenitor cells of HbSS patients with and without proliferative retinopathy. Exp Biol Med 248:677–684. https://doi.org/10.1177/15353702231157927
Banno K, Yoder MC (2018) Tissue regeneration using endothelial colony-forming cells: promising cells for vascular repair. Pediatr Res 83:283–290. https://doi.org/10.1038/pr.2017.231
Flanagan JM, Frohlich DM, Howard TA et al (2011) Genetic predictors for stroke in children with sickle cell anemia. Blood 117:6681–6684. https://doi.org/10.1182/blood-2011-01-332205
Amlie-Lefond C, Flanagan J, Kanter J, Dobyns WB (2018) The genetic landscape of cerebral steno-occlusive arteriopathy and stroke in sickle cell anemia. J Stroke Cerebrovasc Dis 27:2897–2904. https://doi.org/10.1016/j.jstrokecerebrovasdis.2018.06.004
Flanagan JM, Sheehan V, Linder H et al (2013) Genetic mapping and exome sequencing identify 2 mutations associated with stroke protection in pediatric patients with sickle cell anemia. Blood 121:3237–3245. https://doi.org/10.1182/blood-2012-10-464156
Lee J-M, Fernandez-Cadenas I, Lindgren AG (2021) Using human genetics to understand mechanisms in ischemic stroke outcome: from early brain injury to long-term recovery. Stroke 52:3013–3024. https://doi.org/10.1161/STROKEAHA.121.032622
Manzoni C, Kia DA, Vandrovcova J et al (2018) Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief Bioinform 19:286–302. https://doi.org/10.1093/bib/bbw114
Sakamoto TM, Lanaro C, Ozelo MC et al (2013) Increased adhesive and inflammatory properties in blood outgrowth endothelial cells from sickle cell anemia patients. Microvasc Res 90:173–179. https://doi.org/10.1016/j.mvr.2013.10.002
Lin Y, Weisdorf DJ, Solovey A, Hebbel RP (2000) Origins of circulating endothelial cells and endothelial outgrowth from blood. J Clin Investig 105:71–77. https://doi.org/10.1172/JCI8071
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. In: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 15 Oct 2021
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
RStudio Team (2021) RStudio: Integrated Development Environment for R. RStudio, PBC, Boston, MA. https://www.rstudio.com/
Robinson MD, McCarthy DJ, Smyth GK (2010) <tt>edgeR</tt> : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Kolde R (2019) Pheatmap: pretty heatmaps. R package version 1.0.12. In: https://CRAN.Rproject.org/package=pheatmap. Accessed 28 Jul 2023
Blighe K, Lun A (2021) PCAtools: PCAtools: everything principal components analysis. R package version 2.4.0. In: https://github.com/kevinblighe/PCAtools. Accessed 28 Jul 2023
Blighe K, Rana S, Lewis M (2021) EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. R package version 1.10.0. In: https://github.com/kevinblighe/EnhancedVolcano. Accessed 28 Jul 2023
Dennis G, Sherman BT, Hosack DA et al (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3
Doncheva NT, Morris JH, Gorodkin J, Jensen LJ (2019) Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 18:623–632. https://doi.org/10.1021/acs.jproteome.8b00702
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Chin C-H, Chen S-H, Wu H-H et al (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8:S11. https://doi.org/10.1186/1752-0509-8-S4-S11
SankeyMATIC tool SankeyMATIC: a Sankey diagram builder for everyone. In: http://sankeymatic.com/. Accessed 3 Aug 2023
Clarke DJB, Kuleshov MV, Schilder BM et al (2018) eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res 46:W171–W179. https://doi.org/10.1093/nar/gky458
Zhang ZG, Zhang L, Jiang Q, Chopp M (2002) Bone marrow-derived endothelial progenitor cells participate in cerebral neovascularization after focal cerebral ischemia in the adult mouse. Circ Res 90:284–288. https://doi.org/10.1161/hh0302.104460
Li J, Ma Y, Miao X-H et al (2021) Neovascularization and tissue regeneration by endothelial progenitor cells in ischemic stroke. Neurol Sci 42:3585–3593. https://doi.org/10.1007/s10072-021-05428-3
Somanath PR, Razorenova OV, Chen J, Byzova TV (2006) Akt1 in endothelial cell and angiogenesis. Cell Cycle 5:512–518. https://doi.org/10.4161/cc.5.5.2538
Hallstrom TC, Mori S, Nevins JR (2008) An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 13:11–22. https://doi.org/10.1016/j.ccr.2007.11.031
Ladu S, Calvisi DF, Conner EA et al (2008) E2F1 inhibits c-Myc-driven apoptosis via PIK3CA/Akt/mTOR and COX-2 in a mouse model of human liver cancer. Gastroenterology 135:1322–1332. https://doi.org/10.1053/j.gastro.2008.07.012
Chang L, Xi L, Liu Y et al (2018) SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol Med Rep 17:342–349. https://doi.org/10.3892/MMR.2017.7875
Chaussepied M, Ginsberg D (2004) Transcriptional regulation of AKT activation by E2F. Mol Cell 16:831–837. https://doi.org/10.1016/j.molcel.2004.11.003
Meng P, Ghosh R (2014) Transcription addiction: can we garner the Yin and Yang functions of E2F1 for cancer therapy? Cell Death Dis 5(8):e1360–e1360. https://doi.org/10.1038/cddis.2014.326
Nishiyama T, Ladurner R, Schmitz J et al (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143:737–749. https://doi.org/10.1016/j.cell.2010.10.031
Nguyen M-H, Koinuma J, Ueda K et al (2010) Phosphorylation and activation of cell division cycle associated 5 by mitogen-activated protein kinase play a crucial role in human lung carcinogenesis. Cancer Res 70:5337–5347. https://doi.org/10.1158/0008-5472.CAN-09-4372
Xu J, Zhu C, Yu Y et al (2019) Systematic cancer-testis gene expression analysis identified CDCA5 as a potential therapeutic target in esophageal squamous cell carcinoma. EBioMedicine 46:54–65. https://doi.org/10.1016/j.ebiom.2019.07.030
Shen A, Liu L, Chen H et al (2019) Cell division cycle associated 5 promotes colorectal cancer progression by activating the ERK signaling pathway. Oncogenesis 8:19. https://doi.org/10.1038/s41389-019-0123-5
Chen H, Chen J, Zhao L et al (2019) CDCA5, transcribed by E2F1, promotes Oncogenesis by enhancing cell proliferation and inhibiting apoptosis via the AKT pathway in hepatocellular carcinoma. J Cancer 10:1846–1854. https://doi.org/10.7150/jca.28809
Parker LH, Schmidt M, Jin S-W et al (2004) The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature 428:754–758. https://doi.org/10.1038/nature02416
Campagnolo L, Leahy A, Chitnis S et al (2005) EGFL7 is a chemoattractant for endothelial cells and is up-regulated in angiogenesis and arterial injury. Am J Pathol 167:275–284. https://doi.org/10.1016/S0002-9440(10)62972-0
Badiwala MV, Tumiati LC, Joseph JM, et al (2010) Epidermal growth factor-like domain 7 suppresses intercellular adhesion molecule 1 expression in response to hypoxia/reoxygenation injury in human coronary artery endothelial cells. Circulation 122(11 Suppl):S156–61. https://doi.org/10.1161/CIRCULATIONAHA.109.927715
Gustavsson M, Mallard C, Vannucci SJ et al (2007) Vascular response to hypoxic preconditioning in the immature brain. J Cereb Blood Flow Metab 27:928–938. https://doi.org/10.1038/sj.jcbfm.9600408
Li Q, Cheng K, Wang A-Y et al (2019) microRNA-126 inhibits tube formation of HUVECs by interacting with EGFL7 and down-regulating PI3K/AKT signaling pathway. Biomed Pharmacother 116:109007. https://doi.org/10.1016/j.biopha.2019.109007
Schmidt M, Paes K, De Mazière A et al (2007) EGFL7 regulates the collective migration of endothelial cells by restricting their spatial distribution. Development 134:2913–2923. https://doi.org/10.1242/dev.002576
Soncin F (2003) VE-statin, an endothelial repressor of smooth muscle cell migration. EMBO J 22:5700–5711. https://doi.org/10.1093/emboj/cdg549
Wu XY, Liu WT, Wu ZF et al (2016) Identification of HRAS as cancer-promoting gene in gastric carcinoma cell aggressiveness. Am J Cancer Res 6:1935–1948
Li Q, Decker-Rockefeller B, Bajaj A, Pumiglia K (2018) Activation of Ras in the vascular endothelium induces brain vascular malformations and hemorrhagic stroke. Cell Rep 24:2869–2882. https://doi.org/10.1016/j.celrep.2018.08.025
Montaner J, Ramiro L, Simats A et al (2020) Multilevel omics for the discovery of biomarkers and therapeutic targets for stroke. Nat Rev Neurol 16:247–264. https://doi.org/10.1038/s41582-020-0350-6
Pepper MS (1997) Transforming growth factor-beta: vasculogenesis, angiogenesis, and vessel wall integrity. Cytokine Growth Factor Rev 8:21–43. https://doi.org/10.1016/S1359-6101(96)00048-2
Cheng X, Yang Y-L, Li W-H et al (2020) Cerebral ischemia-reperfusion aggravated cerebral infarction injury and possible differential genes identified by RNA-Seq in rats. Brain Res Bull 156:33–42. https://doi.org/10.1016/j.brainresbull.2019.12.014
Downward J (2009) A tumour gene’s fatal flaws. Nature 462:44–45. https://doi.org/10.1038/462044a
Tarassishin L, Suh H-S, Lee SC (2011) Interferon regulatory factor 3 plays an anti-inflammatory role in microglia by activating the PI3K/Akt pathway. J Neuroinflammation 8:187. https://doi.org/10.1186/1742-2094-8-187
Vadas O, Burke JE, Zhang X, et al (2011) Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal 4(195):re2. https://doi.org/10.1126/scisignal.2002165
Thorpe LM, Spangle JM, Ohlson CE et al (2017) PI3K-p110α mediates the oncogenic activity induced by loss of the novel tumor suppressor PI3K-p85α. Proc Natl Acad Sci 114:7095–7100. https://doi.org/10.1073/pnas.1704706114
Chang Milbauer L, Wei P, Enenstein J et al (2008) Genetic endothelial systems biology of sickle stroke risk. Blood 111:3872–3879. https://doi.org/10.1182/blood-2007-06-097188
Zhang SJ, Zhang H, Wei YJ et al (2006) Adult endothelial progenitor cells from human peripheral blood maintain monocyte/macrophage function throughout in vitro culture. Cell Res 16:577–584. https://doi.org/10.1038/sj.cr.7310075
Mikirova NA, Jackson JA, Hunninghake R et al (2009) Circulating endothelial progenitor cells: a new approach to anti-aging medicine? J Transl Med 7:106. https://doi.org/10.1186/1479-5876-7-106
Ding D-C, Shyu W-C, Lin S-Z, Li H (2007) The role of endothelial progenitor cells in ischemic cerebral and heart diseases. Cell Transplant 16:273–284. https://doi.org/10.3727/000000007783464777
Acknowledgements
We thank all the patients; the research staff of Human Genetics Laboratory, Center for Molecular Biology and Genetic Engineering, UNICAMP, Campinas, SP, Brazil and the Hematology and Hemotherapy Center, UNICAMP, Campinas, SP, Brazil.
Funding
This work was supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de S. Paulo) [grant numbers: 2021/14089–0, 2014/00984–3, 2019/18886–1] and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) [finance code 001].
Author information
Authors and Affiliations
Contributions
All authors participated in the design, interpretation of the study and review of the manuscript. Sample collection was performed by Mirta Tomie Ito and Roberta Casagrande Saez; neurological evaluation was conducted by Fernando Cendes; cell isolation and culture were performed by Sueli Matilde da Silva Costa, Mirta Tomie Ito, Roberta Casagrande Saez, Dulcinéia Martins de Albuquerque and Carolina Lanaro; review of medical records was done by Júlia Nicoliello Pereira de Castro, Sueli Matilde da Silva Costa, Victor de Haidar e Bertozzo, Mirta Tomie Ito and Thiago Adalton Rosa Rodrigues; transcriptomic data analysis and interpretation were performed by Júlia Nicoliello Pereira de Castro, Ana Carolina Lima Camargo, Bruno Batista de Souza, Sueli Matilde da Silva Costa and Mônica Barbosa de Melo; gene ontology analysis was conducted by Júlia Nicoliello Pereira de Castro, Sueli Matilde da Silva Costa, Ana Carolina Lima Camargo and Victor de Haidar e Bertozzo; protein–protein interaction network was constructed by Júlia Nicoliello Pereira de Castro and Thiago Adalton Rosa Rodrigues; In silico prediction of regulatory factors was conducted by Júlia Nicoliello Pereira de Castro and Ana Carolina Lima Camargo. The whole work was supervised by Sueli Matilde da Silva Costa, Fernando Ferreira Costa and Mônica Barbosa de Melo. The manuscript was written by Júlia Nicoliello Pereira de Castro, Sueli Matilde da Silva Costa and Mônica Barbosa de Melo. Júlia Nicoliello Pereira de Castro and Sueli Matilde da Silva Costa are equal contributors to the work.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Research Ethics Committees of the Faculty of Medical Sciences, UNICAMP, in accordance with national guidelines (protocol nº 4.859.604) and with the Helsinki Declaration of 1975, as revised in 2008.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
de Castro, J.N.P., da Silva Costa, S.M., Camargo, A.C.L. et al. Comparative transcriptomic analysis of circulating endothelial cells in sickle cell stroke. Ann Hematol 103, 1167–1179 (2024). https://doi.org/10.1007/s00277-024-05655-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-024-05655-6