Abstract
Purpose
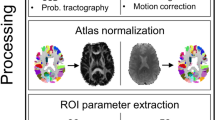
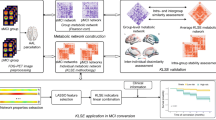
This study evaluates the potential of within-individual Metabolic Connectivity (wi-MC), from dynamic [18F]FDG PET data, based on the Euclidean Similarity method. This approach leverages the biological information of the tracer’s full temporal dynamics, enabling the direct extraction of individual metabolic connectomes. Specifically, the proposed framework, applied to glioma pathology, seeks to assess sensitivity to metabolic dysfunctions in the whole brain, while simultaneously providing further insights into the pathophysiological mechanisms regulating glioma progression.
Methods
We designed an index (Distance from Healthy Group, DfHG) based on the alteration of wi-MC in each patient (n = 44) compared to a healthy reference (from 57 healthy controls), to individually quantify metabolic connectivity abnormalities, resulting in an Impairment Map highlighting significantly compromised areas. We then assessed whether our measure of metabolic network alteration is associated with well-established markers of disease severity (tumor grade and volume, with and without edema). Subsequently, we investigated disruptions in wi-MC homotopic connectivity, assessing both affected and seemingly healthy tissue to deepen the pathology’s impact on neural communication. Finally, we compared network impairments with local metabolic alterations determined from SUVR, a validated diagnostic tool in clinical practice.
Results
Our framework revealed how gliomas cause extensive alterations in the topography of brain networks, even in structurally unaffected regions outside the lesion area, with a significant reduction in connectivity between contralateral homologous regions. High-grade gliomas have a stronger impact on brain networks, and edema plays a mediating role in global metabolic alterations. As compared to the conventional SUVR-based analysis, our approach offers a more holistic view of the disease burden in individual patients, providing interesting additional insights into glioma-related alterations.
Conclusion
Considering our results, individual PET connectivity estimates could hold significant clinical value, potentially allowing the identification of new prognostic factors and personalized treatment in gliomas or other focal pathologies.






Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
Sala A, Lizarraga A, Caminiti SP, et al. Brain connectomics: time for a molecular imaging perspective? Trends Cogn Sci. 2023;27(4):353–66. https://doi.org/10.1016/j.tics.2022.11.015.
Yakushev I, Drzezga A, Habeck C. Metabolic connectivity: methods and applications. Curr Opin Neurol. 2017;30(6):677–85. https://doi.org/10.1097/WCO.0000000000000494.
Veronese M, Moro L, Arcolin M, et al. Covariance statistics and network analysis of brain PET imaging studies. Sci Rep. 2019;9(1). https://doi.org/10.1038/s41598-019-39005-8.
Jamadar SD, Ward PGD, Liang EX, et al. Metabolic and hemodynamic resting-state connectivity of the human brain: a high-temporal resolution simultaneous BOLD-fMRI and FDG-fPET multimodality study. Cereb Cortex. 2021;31(6):2855–67. https://doi.org/10.1093/cercor/bhaa393.
Sun T, Wang Z, Wu Y, et al. Identifying the individual metabolic abnormities from a systemic perspective using whole-body PET imaging. Eur J Nucl Med Mol Imaging. 2022;49(8):2994–3004. https://doi.org/10.1007/s00259-022-05832-7.
Wang M, Jiang J, Yan Z, et al. Individual brain metabolic connectome indicator based on Kullback-Leibler Divergence Similarity Estimation predicts progression from mild cognitive impairment to Alzheimer’s dementia. Eur J Nucl Med Mol Imaging. 2020;47(12):2753–64. https://doi.org/10.1007/s00259-020-04814-x.
Huang SY, Hsu JL, Lin KJ, et al. A Novel Individual Metabolic Brain Network for 18F-FDG PET imaging. Front Neurosci. 2020;14(May):1–11. https://doi.org/10.3389/fnins.2020.00344.
Volpi T, Vallini G, Silvestri E, et al. A new framework for metabolic connectivity mapping using bolus [ 18 F]FDG PET and kinetic modelling. J Cereb Blood Flow Metab. 2022;1–21. https://doi.org/10.1177/0271678X231184365.
Liptrot M, Adams KH, Martiny L, et al. Cluster analysis in kinetic modelling of the brain: a noninvasive alternative to arterial sampling. NeuroImage. 2004;21(2):483–93. https://doi.org/10.1016/j.neuroimage.2003.09.058.
Bertoldo A, Rizzo G, Veronese M. Deriving physiological information from PET images: From SUV to compartmental modelling [Internet]. Vol. 2, Clinical and Translational Imaging. Springer-Verlag Italia s.r.l.; 2014. 239–51. https://doi.org/10.1007/s40336-014-0067-x.
Mancusi R, Monje M. The neuroscience of cancer. Nature. 2023;618(7965):467–79. https://doi.org/10.1038/s41586-023-05968-y.
Daniel AGS, Hacker CD, Lee JJ, et al. Homotopic functional connectivity disruptions in glioma patients are associated with tumor malignancy and overall survival. Neuro-Oncology Adv. 2021;3(1):1–10. https://doi.org/10.1093/noajnl/vdab176.
Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–20. https://doi.org/10.1007/s00401-016-1545-1.
Goyal MS, Blazey T, Metcalf NV, et al. Brain aerobic glycolysis and resilience in Alzheimer disease. Proc Natl Acad Sci U S A. 2023;120(7):1–8. https://doi.org/10.1073/pnas.2212256120.
Hammers A, Allom R, Koepp MJ, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19(4):224–47. https://doi.org/10.1002/hbm.10123.
Yan X, Kong R, Xue A, et al. Homotopic local-global parcellation of the human cerebral cortex from resting-state functional connectivity. NeuroImage. 2023;273(October 2022):120010. https://doi.org/10.1016/j.neuroimage.2023.120010.
Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54(3):2033–44. https://doi.org/10.1016/j.neuroimage.2010.09.025.
Silvestri E, Moretto M, Facchini S, et al. Widespread cortical functional disconnection in gliomas: an individual network mapping approach. Brain Commun. 2022;4(2):1–14. https://doi.org/10.1093/braincomms/fcac082.
Andersen SM, Rapcsak SZ, Beeson PM. Cost function masking during normalization of brains with focal lesions: still a necessity? NeuroImage. 2010;53(1):78–84. https://doi.org/10.1016/j.neuroimage.2010.06.003.
Feng X, Deistung A, Dwyer MG, et al. An improved FSL-FIRST pipeline for subcortical gray matter segmentation to study abnormal brain anatomy using quantitative susceptibility mapping (QSM). Magn Reson Imaging. 2017;39:110–22. https://doi.org/10.1016/j.mri.2017.02.002.
Deng S, Franklin CG, O’Boyle M, et al. Hemodynamic and metabolic correspondence of resting-state voxel-based physiological metrics in healthy adults. NeuroImage. 2022;250(January):118923. https://doi.org/10.1016/j.neuroimage.2022.118923.
Nugent S, Croteau E, Potvin O, et al. Selection of the optimal intensity normalization region for FDG-PET studies of normal aging and Alzheimer’s disease. Sci Rep. 2020;10(1):1–8. https://doi.org/10.1038/s41598-020-65957-3.
Miller KD, Ostrom QT, Kruchko C, et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J Clin. 2021;71(5):381–406. https://doi.org/10.3322/caac.21693.
Daniel AGS, Park KY, Roland JL, et al. Functional connectivity within glioblastoma impacts overall survival. Neuro Oncol. 2021;23(3):412–21. https://doi.org/10.1093/neuonc/noaa189.
Horwitz B, Duara R, Rapoport SI. Intercorrelations of glucosemetabolic rates between brain regions: application to healthy males in a state of reduced sensory input. J Cereb Blood Flow Metab. 1984;4(4):484–99. https://doi.org/10.1038/jcbfm.1984.73.
Feeney DM, Baron JC, Diaschisis. Stroke. 1986;17(5):817–30. https://doi.org/10.1161/01.STR.17.5.817.
Jung E, Alfonso J, Monyer H, et al. Neuronal signatures in cancer. Int J Cancer. 2020;147(12):3281–91. https://doi.org/10.1002/ijc.33138.
Lv K, Hu Y, Cao X, et al. Altered whole-brain functional network in patients with frontal low-grade gliomas: a resting-state functional MRI study. Neuroradiology. 2024;66(5):775–84. https://doi.org/10.1007/s00234-024-03300-7.
Lin ZX. Glioma-related edema: New insight into molecular mechanisms and their clinical implications. Chin J Cancer. 2013;32(1):49–52. https://doi.org/10.5732/cjc.012.10242.
Karavasilis E, Christidi F, Velonakis G, et al. Ipsilateral and contralateral cerebro-cerebellar white matter connections: a diffusion tensor imaging study in healthy adults. J Neuroradiol. 2019;46(1):52–60. https://doi.org/10.1016/j.neurad.2018.07.004.
Buckner RL, Krienen FM, Castellanos A, et al. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(5):2322–45. https://doi.org/10.1152/jn.00339.2011.
Nenning KH, Furtner J, Kiesel B, et al. Distributed changes of the functional connectome in patients with glioblastoma. Sci Rep. 2020;10(1):1–11. https://doi.org/10.1038/s41598-020-74726-1.
Baumann C, Tichy J, Schaefer JH, et al. Delay in diagnosing patients with right-sided glioblastoma induced by hemispheric-specific clinical presentation. J Neurooncol. 2020;146(1):63–9. https://doi.org/10.1007/s11060-019-03335-4.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Funding for this research was provided by the McDonnell Center for Systems Neuroscience and the NIH/ NIA R01AG053503 (AGV) and R01AG057536 (AGV, MSG). Some of the MRI sequences used were obtained from the Massachusetts General Hospital.
Author information
Authors and Affiliations
Contributions
Andrei G. Vlassenko and Diego Cecchin collected the data. Giulia Vallini and Alessandra Bertoldo designed the research. Giulia Vallini, Erica Silvestri and Tommaso Volpi analyzed the data. Giulia Vallini, Erica Silvestri, Tommaso Volpi, John J. Lee, Andrei G. Vlassenko, Manu S. Goyal, Diego Cecchin, Maurizio Corbetta and Alessandra Bertoldo interpreted the results. Giulia Vallini wrote the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Ethical approval
All assessments and imaging procedures were approved by Human Research Protection Office and Radioactive Drug Research Committee at Washington University in St. Louis (healthy controls). Concerning glioma patients, the protocol has been approved by the local Ethics Committee of the University Hospital of Padova. All procedures performed in studies were conducted in accordance with the 1964 Declaration of Helsinki and its subsequent amendments.
Consent to participate
Informed written consent was obtained from all individual participants included in the study.
Consent to publish
Participants signed informed consent regarding publishing their data.
Competing interests
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vallini, G., Silvestri, E., Volpi, T. et al. Individual-level metabolic connectivity from dynamic [18F]FDG PET reveals glioma-induced impairments in brain architecture and offers novel insights beyond the SUVR clinical standard. Eur J Nucl Med Mol Imaging 52, 836–850 (2025). https://doi.org/10.1007/s00259-024-06956-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-024-06956-8