Abstract
Purpose
To evaluate diagnostic accuracy of fully automated analysis of multimodal imaging data using [18F]-FET-PET and MRI (including amide proton transfer-weighted (APTw) imaging and dynamic-susceptibility-contrast (DSC) perfusion) in differentiation of tumor progression from treatment-related changes in patients with glioma.
Material and methods
At suspected tumor progression, MRI and [18F]-FET-PET data as part of a retrospective analysis of an observational cohort of 66 patients/74 scans (51 glioblastoma and 23 lower-grade-glioma, 8 patients included at two different time points) were automatically segmented into necrosis, FLAIR-hyperintense, and contrast-enhancing areas using an ensemble of deep learning algorithms. In parallel, previous MR exam was processed in a similar way to subtract preexisting tumor areas and focus on progressive tumor only. Within these progressive areas, intensity statistics were automatically extracted from [18F]-FET-PET, APTw, and DSC-derived cerebral-blood-volume (CBV) maps and used to train a Random Forest classifier with threefold cross-validation. To evaluate contribution of the imaging modalities to the classifier’s performance, impurity-based importance measures were collected. Classifier performance was compared with radiology reports and interdisciplinary tumor board assessments.
Results
In 57/74 cases (77%), tumor progression was confirmed histopathologically (39 cases) or via follow-up imaging (18 cases), while remaining 17 cases were diagnosed as treatment-related changes. The classification accuracy of the Random Forest classifier was 0.86, 95% CI 0.77–0.93 (sensitivity 0.91, 95% CI 0.81–0.97; specificity 0.71, 95% CI 0.44–0.9), significantly above the no-information rate of 0.77 (p = 0.03), and higher compared to an accuracy of 0.82 for MRI (95% CI 0.72–0.9), 0.81 for [18F]-FET-PET (95% CI 0.7–0.89), and 0.81 for expert consensus (95% CI 0.7–0.89), although these differences were not statistically significant (p > 0.1 for all comparisons, McNemar test). [18F]-FET-PET hot-spot volume was single-most important variable, with relevant contribution from all imaging modalities.
Conclusion
Automated, joint image analysis of [18F]-FET-PET and advanced MR imaging techniques APTw and DSC perfusion is a promising tool for objective response assessment in gliomas.
Similar content being viewed by others
Introduction
Managing glioma patients, radiologists and clinicians often need to distinguish progressive disease (PD) from treatment-related changes (TRCs). Radiologic assessment of tumor response and progression is traditionally based on volumetric changes of the enhancing tumor area. The discrimination between PD and TRC (such as blood–brain-barrier breakdown following radiotherapy) is however extremely challenging as both typically present with new or progressive contrast enhancement (CE) on T1-weighted gadolinium-enhanced magnetic resonance imaging (MRI) [1,2,3]. While for PD, increased contrast enhancement is typically the result of neoangiogenesis, changes in contrast enhancement after therapy can result from a variety of nontumorous processes, such as ischemia, postsurgical changes, treatment-related inflammation, subacute radiation effects, and radiation necrosis [4].
Diagnosis and treatment of PD and TRC requires multidisciplinary structures of care, and defined processes. Diagnosis has to be made on an interdisciplinary level with the joint knowledge of a neuroradiologist, radiation oncologist, neurosurgeon, and neurooncologist. A multi-step approach as an opportunity to review as many characteristics as possible to improve diagnostic confidence is recommended. Additional information about radiotherapy (RT) techniques are crucial for diagnosis. Yet, pathologic confirmation is considered the most reliable method to differentiate PD from TRC. However, to avoid unnecessary surgery, many efforts have been undertaken to improve non-invasive tumor response assessment. In view of the inability of traditional MRI including T1-weighted or T2-weighted sequences to reliably differentiate PD and TRC [5], advanced imaging modalities aiming to visualize tumor biology and key oncogenic processes have been frequently studied [6].
Relative cerebral blood volume (rCBV) obtained from MR-based dynamic susceptibility contrast (DSC) perfusion imaging has been histopathologically confirmed to provide evidence of neoangiogenesis [7] — a hallmark of malignant gliomas. Some studies have reported that median rCBV and histogram analysis of rCBV can help to differentiate TRC from PD [8, 9] by distinguishing between vital tumor tissue in PD and other causes of contrast enhancement in TRC.
A relatively novel but promising molecular MRI technique is amide proton transfer-weighted (APTw) imaging. This imaging method capitalizes on the constant dissociation and transfer of amide-bound hydrogen atoms into the surrounding water. Previous studies proofed the benefit of APTw imaging in the differentiation between the different WHO grades of gliomas [10] as well as the ability to differentiate between tumor progression and therapy-related changes [11, 12].
With FET as a tracer, PET is known to visualize the amino acid uptake in gliomas and thus metabolically active tumor cells [13]. Several studies exist that demonstrated the clinical utility of [18F]-FET-PET for preoperative grading [14] and biopsy planning [15] as well as for differentiating tumor progression from treatment-related changes [16].
Combining information from different techniques and imaging modalities can help to further decipher the complex diagnosis of progress and TRC in gliomas. A few studies have already investigated the added value of APTw to diffusion- and perfusion-weighted imaging [17] or methionine PET [18]. However, traditional assessment of multimodal imaging data is challenging: Extracting (semi-)quantitative information (such as tumor-to-background ratio (TBR)) from such images is observer-dependent despite means to better standardize this. Further, truly integrating imaging information from multiple modalities (in a region- or even voxel-wise fashion) requires high proficiency in reading these images [19].
In view of recent advances in deep learning for medical image analysis, we developed a fully automated pipeline for longitudinal assessment of changes in tumor morphology, unbiased extraction of quantitative imaging information in these areas from multimodal, advanced imaging modalities, and ultimately predictive modeling of response assessment. Here, we evaluated the accuracy of such a model-based differentiation between PD and TRC [20].
Material and methods
Patient data
All patients were part of a consecutive, prospective observational glioma cohort from December 2017 to April 2020, approved by our local Institutional Review Board. All patients gave written informed consent.
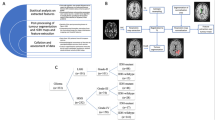
For this retrospective study, we analyzed data of 66 patients (74 MRI and [18F]-FET-PET scans) with histologically confirmed glioma (WHO grades I–IV) according to the 2016 WHO classification of CNS tumors [21]. We included all patients in this time frame who (a) had available MR imaging (including T2- and T1-weighted imaging before and after administration of gadolinium-based contrast agent plus APTw imaging and DSC perfusion) and who (b) had suspected tumor progression with consecutively performed [18F]-FET-PET, (c) with interdisciplinary consensus reading in our neurooncology tumor board, and (d) where either histopathological or additional long-term follow-up confirmation of the diagnosis (PD or TRC) was available (Fig. 1). All images were rated according to the RANO criteria [22], and cases with a morphologically mixed appearance in MRI and/or PET scan (containing both areas with PD and TRC) were accordingly rated as progressive disease (MRI 4.1%; 3/74 and [18F]-FET-PET 8.1%; 6/74). MRI and PET assessment were taken from the clinical report. Eight patients were included twice and analyzed as separate cases as they fulfilled inclusion criteria at two different points of time (between the two included scans, four of them underwent resection, three chemotherapy, and one radio-chemotherapy). Further patient characteristics are given in Table 1.
Image acquisition
[18F]-FET-PET data were acquired on a PET/MR (Biograph mMR, Siemens Healthcare GmbH, Erlangen, Germany), n = 57, and a PET/CT (Biograph mCT; Siemens Healthcare, Knoxville, TN, USA), n = 24, according to a standard clinical protocol. Patients were asked to fast for a minimum of 4 h before scanning. Emission scans were acquired at 30 to 40 min after intravenous injection of a target dose of 185 ± 10% MBq [18F]-FET. Attenuation correction was performed according to vendor’s protocol.
The MR imaging was performed on a Philips (Best, the Netherlands) 3 T scanner (Achieva or Ingenia). Our MR protocol included an isotropic FLAIR (voxel size 1 mm3, TE = 269 ms, TR = 4800 ms, TI = 1650 ms), isotropic T1-TFE (voxel size 1 mm3, TE = 4 ms, TR = 9 ms) before and after contrast, axial T2 (voxel size 0.36 × 0.36 × 4 mm3, TE = 87 ms, TR = 3396 ms), 3D APTw (fast spin echo, voxel size 0.9 × 0.9 × 1.8 mm3, TE = 7.8 ms, TR = 6 s, RF saturation pulse train B1,rms = 2 μT, Tsat = 2 s, duty-cycle 100%, 9 volumes ω = ± 3.5 ppm ± 0.8 ppm and reference ω0 = − 1560 ppm, intrinsic B0 correction [23], MTR asymmetry at + 3.5 ppm as APT-weighted = APTw contrast), as well as DSC perfusion (voxel size 1.75 × 1.75 × 4 mm3, TE = 40 ms, TR = 1547 ms, Flip Angle = 75°, 80 dynamics).
Image processing
Processing of DSC data for rCBV parameter maps used custom programs in MATLAB R2019b (MathWorks, Natick, MA, USA). Spatial coregistration of the different modalities and segmentation of anatomical images for gray matter (GM), white matter (WM), and CSF were conducted using SPM12 (www.fil.ion.ucl.ac.uk/spm) with standard parameter settings. Leakage‐corrected CBV values were obtained using a reference curve approach and numerical integration [24,25,26]. Relative CBV (rCBV) values were calculated by assuming healthy WM values of 2.5% [27].
Post-processing of APTw images followed the vendor’s standard implementation.
Image analysis
All images and parameter maps ([18F]-FET-PET, CBV, APTw) from a single patient were spatially normalized into the SRI24 atlas space [28] and resampled to 1 mm isotropic resolution using a rigid, mutual information-driven registration with the open-source ANTs software (https://stnava.github.io/ANTs/) [29]. Tumors were automatically segmented into necrosis, contrast-enhancing tumor, and FLAIR-hyperintense tumor, using the freely available BraTS Toolkit developed by us [30]. In brief, BraTS Toolkit ensembles several brain tumor segmentation algorithms, relying on a multimodal input of T1w, T1w with contrast, T2, and FLAIR images, and fuses the resulting candidate segmentations into a final consensus segmentation using SIMPLE fusion [31]. All registrations and segmentations were checked and — where necessary — corrected manually by KP (board-certified radiologist with 8 years of experience).
To limit subsequent image analysis to tumor regions with progressive signal alterations, we subtracted the tumor segmentation of the previous exam from the current segmentation and excluded necrotic areas.
From these segmentation areas, and using the coregistered sequences, we extracted pre-defined summary statistics (5th, 25th, 50th, 75th, and 95th percentile intensity, inter-quartile range, and Shannon entropy) as well as volumes of hot-spot areas delineated by the different modalities using a Python script. For hot-spot definition, we relied on pre-defined thresholds from the literature by using the following cut offs: APTw > 1.79 [12], tumor-background ratio > 2 for [18F]-FET-PET [32], and rCBV > 5.6 [33]. For estimation of the tumor-background ratio for the [18F]-FET-PET data, we relied on the white matter maps generated during CBV processing, excluding tumor areas, to calculate the background signal.
This feature vector was used as input for a Random Forest classifier [34], an ensemble-based machine learning algorithm which is known for its relative resistance to over-fitting as well as its ability to deal with correlated data. We used the scikit-learn implementation, leaving all parameters at their standard recommended default values, with a threefold cross-validation for obtaining unbiased estimates of the classifier performance. We also collected impurity-based feature importance to judge the contribution of the imaging modalities to the classifier’s performance.
Statistical analysis
A receiver operator curve analysis was performed for the classifier based on all imaging data. Results are from a threefold cross-validation. Based on a cut off value of 0.5, we plotted confusion matrices and calculated sensitivity, specificity, and accuracy. Differences between the binary accuracy and the no-information rate in our data set were tested using a binomial test, with the no-information rate as success probability. Prediction accuracy between the different models were compared using a McNemar test.
Results
Sixty-six patients (seventy-four cases) met our inclusion criteria. They had a median age of 55 years (range 54.91 ± 12.2), 45% of them were female (Table 1). Median interval between [18F]-FET-PET and MRI was 18.2 days. The reference standard used in this study, i.e., the final diagnosis of true progressive disease (PD, n = 57) and TRC (n = 17), was based on histopathology in 43 cases (resection or biopsy, n = 39 PD and n = 4 TRC) and on interdisciplinary board consensus based on follow-up MRI imaging (including APTw and CBV) after 3 months in 31 cases (n = 18 PD, n = 13 TRC) and according to the RANO criteria. In MRI, progression was suspected in 64 of 74 cases, whereas in [18F]-FET-PET, real progression was suspected in only 49/74 cases. In contrast, TRC was correctly identified only in 7/17 cases in MRI, but in 14/17 in PET.
ROC analysis for the identification of PD in our fully automated data analysis (results from a threefold cross-validation) yielded an area under the curve (AUC) of 0.85 (Fig. 2) with an accuracy of 0.86 (sensitivity 0.91, specificity 0.701). This model performance was significantly greater than the no-information rate in our data set (i.e., the rate of patients with PD; p = 0.03). Compared with these results, traditional assessment, relying either on MRI (accuracy: 0.82; sensitivity: 0.95; specificity: 0.41) or [18F]-FET-PET (accuracy: 0.81; sensitivity: 0.81; specificity: 0.82) as well as the interdisciplinary tumor board expert consensus (accuracy: 0.81; sensitivity: 0.81; specificity: 0.53), showed a lower diagnostic accuracy not significantly above chance (Table 2). Upon comparison of the machine learning model with predictions based on either MRI (p = 0.774) or [18F]-FET-PET (p = 0.424), we noted no significant differences.
Upon inspection of the final model and the individual contribution of imaging features (Fig. 3), imaging information derived from [18F]-FET-PET data contributed most importantly to the classifier. However, also features from APTw and CBV maps ranked high, indicating a reliance of the classifier on joint imaging information (Figs. 4 and 5).
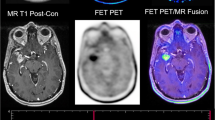
Example images of progressive disease in a 65-year-old female patient with left frontal GBM and a new contrast enhancement superior to the former resection area (upper left: ce T1-w, lower left: previous time point, upper middle: automated segmentation overlay (green: new FLAIR edema area; yellow: new ce area), lower middle: CBV, upper right: [18F]-FET-PET, lower right APTw)
Example images of TRC in a 34-year-old male with a new contrast enhancing focus next to the resection cavity after therapy of a left frontal GBM (upper left: ce T1-w, lower left: previous time point, upper middle: automated segmentation overlay (green: new FLAIR edema area; yellow: new ce area), lower middle: CBV, upper right: [18F]-FET-PET, lower right APTw)
Discussion
To allow for objective, user-independent assessment of therapy response, we have developed an automated, joint image analysis of [18F]-FET-PET and advanced MR imaging techniques. The classification accuracy of the Random Forest classifier was 0.86 (sensitivity 0.91, specificity 0.71) and therefore significantly above the no-information rate of 0.77 (p = 0.03) compared to an accuracy of 0.82 for MRI, 0.81 for [18F]-FET-PET, and 0.81 for expert consensus. The single-most important variable was [18F]-FET-PET hot-spot volume, with relevant contribution from all imaging modalities.
Diagnosis and in particular response assessment of brain tumors is strongly based on imaging, especially MRI techniques [35,36,37], because histological confirmation often cannot be realized easily and bears substantial risks. In the future, artificial intelligence (AI) has the potential to improve image-based diagnosis [38] and augment clinical decision-making in the management of oncologic patients [39]. However, assessing the rich, multimodal imaging information is a complex task, and in particular when MR and [18F]-FET-PET [40, 41] images need to be assessed jointly, requires expertise from multiple disciplines. In many institutions, interdisciplinary analysis of these images (and the patient history) in tumor boards has therefore become standard of care. Even there however, the extraction of (semi-)quantitative information from these images (such as tumor-background ratio from [18F]-FET-PET images [42]) is limited by its inherent inter-observer variability and the tediousness of manually delineating (progressive) tumor areas on images. Promising to overcome these challenges, advances in automated image analysis offer the perspective of objective, multimodal assessment of tumor response [43, 44]. As highlighted in the Brain Tumor Segmentation Challenge (BraTS) [45], machine learning-based glioma segmentation has now reached a point where the algorithms’ performance is non-inferior to human raters. This has been shown to facilitate objective response assessment according to the RANO criteria in a large, prospective trial cohort [38]. In parallel, machine learning algorithms excel at the (non-linear) analysis of multimodal data and thus offer a means to harvest the wealth of information contained in medical imaging data [20, 43].
Assessing the synergistic value of multiple imaging techniques for glioma diagnosis and response assessment has been investigated by several groups. Park et al. [46] for example concluded that adding APT imaging to conventional and perfusion MRI significantly improves the diagnostic performance for differentiating true progression from pseudoprogression. Furthermore, they found that a combination of contrast-enhanced T1w, nCBV90, and APT90 resulted in greater diagnostic accuracy for differentiating true progression from pseudoprogression than the combination of contrast-enhanced T1w and nCBV90 alone. Liesche et al. [47] showed that 18F-fluoroethyl-tyrosine uptake is correlated with amino acid transport and neovascularization in treatment-naive glioblastomas. Following these results, a study by Schön et al. [48] compared the synergism of amino acid PET, amide proton transfer, and perfusion-weighted MRI in newly diagnosed gliomas. They found that the overlap between APTw/CBV is relevantly lower than between APTw/FET, indicating a potential synergistic value of combining APTw and CBV information. This was further corroborated by investigating the stereotactic biopsies, where they found a more pronounced association of CBV and vascularity (compared with APTw) on the one side, and a stronger correlation of APTw with cellularity on the other side. Their findings also underlined the future diagnostic potential of a multimodal imaging concept in making oncogenic processes visible and supporting decision-making in clinically challenging situations such as grading gliomas or differentiating radiation necrosis from real progression. Along this line, such information-rich data sets serve as an ideal basis for training machine learning classifiers which are able to (non-linearly) integrate the multimodal input data, as we and others have previously demonstrated [49]. Inspecting the final model and individual contribution of imaging features in our study, imaging information derived from [18F]-FET-PET data contributed most importantly to the classifier (Fig. 3). This predominance of [18F]-FET-PET information has been suggested by others [50]. However, also features from APTw and CBV maps ranked high, indicating a reliance of the classifier on joint imaging information. The importance of integrating multimodal information is also highlighted by the results for our algorithm compared with the individual assessment of 18F]-FET-PET and MRI (Fig. 2, Table 2): While in MRI, there was a bias towards diagnosing PD (10 cases of TRC mis-diagnosed as PD), whereas in [18F]-FET-PET, the opposite was the case (11 cases of PD mis-diagnosed as TRC). This was balanced out in the multimodal Random Forest classifier (5 vs. 5) and — to a lesser degree — also in the expert consensus including both departments of neuroradiology and nuclear medicine (8 vs. 6).
Up to now, only few studies are available that involve machine learning techniques to differentiate between treatment-related changes and real tumor progression. An SVM (support vector machine) classifier has been trained to diagnose pseudoprogression vs. recurrence in patients with glioma treated with surgery and chemotherapy. In their study containing 31 patients, Hu et al. showed a sensitivity and specificity of the classifier for pseudoprogression of 89.91 and 93.72%, respectively, with AUC of 0.94; with DWI and rCBV as the best predictor image sequences [51]. A CNN has been developed to differentiate true progressive disease from pseudoprogression in patients with a GBM status post chemo-radiation and surgery with a performance of AUC = 0.83, comparable to our MR-only results [3]. However, these studies still required manual labeling of tumor regions (or in the case of the study by Jang et al. the selection of tumor-bearing slices) for subsequent analysis. In contrast, our method is fully automated, from longitudinal tumor segmentation and feature extraction to classification.
In our study, we decided not to limit our analysis to patients with suspected pseudoprogression (i.e., those with new or progressive contrast enhancement following radiotherapy) but rather include all patients where response assessment proved difficult enough to warrant [18F]-FET-PET imaging and discussion in our interdisciplinary tumor board. Despite these broad inclusion criteria, 23% of patients showed TRC; a number in line with the literature [1]. Further, we have included eight patients twice. While this might induce a bias in our model, this choice reflects real clinical practice potentially better than opposed to strict inclusion criteria, which tend to limit generalizability of results and possibly bias models towards patients meeting these strict inclusion criteria. Importantly though, these eight patients had a change of treatment between both time points included in our study and therefore were not included twice for the same therapy.
As our results are promising, the present study has some limitations. First of all, we used a unicentric study as well as a relatively small sample size. In particular, our results were only validated on internal data using a cross-validation approach. The lack of an (ideally external) test set as therefore to be considered when interpreting our results. How readily our models generalize to data from other hospitals/scanners if therefore also not clear. We opted for an advanced imaging protocol — including APTw imaging — which might limit the immediate broad applicability of our findings. On the other hand, our approach thus highlights the value of multimodal imaging, capturing different aspects of tumor biology, and image analysis. When excluding APTw imaging, we observe a model AUC of 0.82. Future studies will be necessary to determine the best combination of imaging modalities; however, upon inspection of feature importance, we also find relevant information from APTw. We included FET-PET data from both a PET CT scanner as well as a PET MR scanner. While this improves the variability in our data and potentially also generalizability, we did not assess the influence of the differences in attenuation correction. For modeling our data, we chose a Random Forest, which is a well-established machine learning model for classification in the presence of (potentially) correlated input data. While modern deep learning approaches are able to learn even more complex decision boundaries and might potentially outperform a Random Forest, we were lacking enough data to train (and in particular independently test) such a model. Also, in particular, the segmentation ensemble necessitates the presence of additional hardware (GPU). Integration of such algorithms into clinical routine is a further challenge to be solved. While histology is considered gold standard for response assessment, tumor progression and TRC were determined by follow-up imaging in some cases and could not be validated by histological data. Finally, as most of the patients diagnosed with PD previously underwent radiotherapy, histopathologic samples of real progression in reality often show a mixture of areas of true progression as well as (microscopic) therapy-related changes, a fact neither MRI nor [18F]-FET-PET is able to resolve up to now. As tumor heterogeneity is a challenging fact, further studies — especially with the help of automated response assessments as a promising tool — have to be conducted to improve the detection and resolution of local tumor heterogeneity.
Conclusions
Predicting tumor biology and response on imaging using AI is promising to play an important role in future practice. Our study shows that [18F]-FET-PET, multiparametric MRI with APTw and DSC perfusion parameters can be combined in a fully automated analysis to help objectively evaluate treatment response in gliomas and may therefore aid in the optimal care of these patients. The promise and performance of AI techniques in daily clinical practice and their effect on patient outcomes warrant further development.
Data availability
The data sets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
N/A
References
Brandsma D, Stalpers L, Taal W, Sminia P, van den Bent MJ. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008;9:453–61. https://doi.org/10.1016/S1470-2045(08)70125-6.
Topkan E, Topuk S, Oymak E, Parlak C, Pehlivan B. Pseudoprogression in patients with glioblastoma multiforme after concurrent radiotherapy and temozolomide. Am J Clin Oncol. 2012;35:284–9. https://doi.org/10.1097/COC.0b013e318210f54a.
Jang BS, Jeon SH, Kim IH, Kim IA. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci Rep. 2018;8:12516. https://doi.org/10.1038/s41598-018-31007-2.
Chu HH, Choi SH, Ryoo I, Kim SC, Yeom JA, Shin H, et al. Differentiation of true progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide: comparison study of standard and high-b-value diffusion-weighted imaging. Radiology. 2013;269:831–40. https://doi.org/10.1148/radiol.13122024.
Abdulla S, Saada J, Johnson G, Jefferies S, Ajithkumar T. Tumour progression or pseudoprogression? A review of post-treatment radiological appearances of glioblastoma. Clin Radiol. 2015;70:1299–312. https://doi.org/10.1016/j.crad.2015.06.096.
Le Fevre C, Constans JM, Chambrelant I, Antoni D, Bund C, Leroy-Freschini B, et al. Pseudoprogression versus true progression in glioblastoma patients: a multiapproach literature review. Part 2 - Radiological features and metric markers. Crit Rev Oncol Hematol. 2021;159:103230. https://doi.org/10.1016/j.critrevonc.2021.103230.
Jain R, Gutierrez J, Narang J, Scarpace L, Schultz LR, Lemke N, et al. In vivo correlation of tumor blood volume and permeability with histologic and molecular angiogenic markers in gliomas. AJNR Am J Neuroradiol. 2011;32:388–94. https://doi.org/10.3174/ajnr.A2280.
Kim HS, Kim JH, Kim SH, Cho KG, Kim SY. Posttreatment high-grade glioma: usefulness of peak height position with semiquantitative MR perfusion histogram analysis in an entire contrast-enhanced lesion for predicting volume fraction of recurrence. Radiology. 2010;256:906–15. https://doi.org/10.1148/radiol.10091461.
Mangla R, Singh G, Ziegelitz D, Milano MT, Korones DN, Zhong J, et al. Changes in relative cerebral blood volume 1 month after radiation-temozolomide therapy can help predict overall survival in patients with glioblastoma. Radiology. 2010;256:575–84. https://doi.org/10.1148/radiol.10091440.
Choi YS, Ahn SS, Lee SK, Chang JH, Kang SG, Kim SH, et al. Amide proton transfer imaging to discriminate between low- and high-grade gliomas: added value to apparent diffusion coefficient and relative cerebral blood volume. Eur Radiol. 2017;27:3181–9. https://doi.org/10.1007/s00330-017-4732-0.
Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med. 2011;17:130–4. https://doi.org/10.1038/nm.2268.
Jiang S, Eberhart CG, Lim M, Heo HY, Zhang Y, Blair L, et al. Identifying recurrent malignant glioma after treatment using amide proton transfer-weighted MR imaging: a validation study with image-guided stereotactic biopsy. Clin Cancer Res. 2019;25:552–61. https://doi.org/10.1158/1078-0432.CCR-18-1233.
Heiss P, Mayer S, Herz M, Wester HJ, Schwaiger M, Senekowitsch-Schmidtke R. Investigation of transport mechanism and uptake kinetics of O-(2-[18F]fluoroethyl)-L-tyrosine in vitro and in vivo. J Nucl Med. 1999;40:1367–73.
Jansen NL, Schwartz C, Graute V, Eigenbrod S, Lutz J, Egensperger R, et al. Prediction of oligodendroglial histology and LOH 1p/19q using dynamic [(18)F]FET-PET imaging in intracranial WHO grade II and III gliomas. Neuro Oncol. 2012;14:1473–80. https://doi.org/10.1093/neuonc/nos259.
Kunz M, Thon N, Eigenbrod S, Hartmann C, Egensperger R, Herms J, et al. Hot spots in dynamic (18)FET-PET delineate malignant tumor parts within suspected WHO grade II gliomas. Neuro Oncol. 2011;13:307–16. https://doi.org/10.1093/neuonc/noq196.
Galldiks N, Dunkl V, Stoffels G, Hutterer M, Rapp M, Sabel M, et al. Diagnosis of pseudoprogression in patients with glioblastoma using O-(2-[18F]fluoroethyl)-L-tyrosine PET. Eur J Nucl Med Mol Imaging. 2015;42:685–95. https://doi.org/10.1007/s00259-014-2959-4.
Togao O, Hiwatashi A, Yamashita K, Kikuchi K, Keupp J, Yoshimoto K, et al. Grading diffuse gliomas without intense contrast enhancement by amide proton transfer MR imaging: comparisons with diffusion- and perfusion-weighted imaging. Eur Radiol. 2017;27:578–88. https://doi.org/10.1007/s00330-016-4328-0.
Park JE, Lee JY, Kim HS, Oh JY, Jung SC, Kim SJ, et al. Amide proton transfer imaging seems to provide higher diagnostic performance in post-treatment high-grade gliomas than methionine positron emission tomography. Eur Radiol. 2018;28:3285–95. https://doi.org/10.1007/s00330-018-5341-2.
Unterrainer M, Vettermann F, Brendel M, Holzgreve A, Lifschitz M, Zahringer M, et al. Towards standardization of (18)F-FET PET imaging: do we need a consistent method of background activity assessment? EJNMMI Res. 2017;7:48. https://doi.org/10.1186/s13550-017-0295-y.
Wiestler B, Menze B. Deep learning for medical image analysis: a brief introduction. Neurooncol Adv. 2020;2:iv35-iv41. https://doi.org/10.1093/noajnl/vdaa092.
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–20. https://doi.org/10.1007/s00401-016-1545-1.
Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–72. https://doi.org/10.1200/JCO.2009.26.3541.
Togao O, Keupp J, Hiwatashi A, Yamashita K, Kikuchi K, Yoneyama M, et al. Amide proton transfer imaging of brain tumors using a self-corrected 3D fast spin-echo dixon method: comparison with separate B0 correction. Magn Reson Med. 2017;77:2272–9. https://doi.org/10.1002/mrm.26322.
Kluge A, Lukas M, Toth V, Pyka T, Zimmer C, Preibisch C. Analysis of three leakage-correction methods for DSC-based measurement of relative cerebral blood volume with respect to heterogeneity in human gliomas. Magn Reson Imaging. 2016;34:410–21. https://doi.org/10.1016/j.mri.2015.12.015.
Hedderich D, Kluge A, Pyka T, Zimmer C, Kirschke JS, Wiestler B, et al. Consistency of normalized cerebral blood volume values in glioblastoma using different leakage correction algorithms on dynamic susceptibility contrast magnetic resonance imaging data without and with preload. J Neuroradiol. 2019;46:44–51. https://doi.org/10.1016/j.neurad.2018.04.006.
Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27:859–67.
Leenders KL. PET: blood flow and oxygen consumption in brain tumors. J Neurooncol. 1994;22:269–73. https://doi.org/10.1007/BF01052932.
Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp. 2010;31:798–819. https://doi.org/10.1002/hbm.20906.
Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–44. https://doi.org/10.1016/j.neuroimage.2010.09.025.
Kofler F, Berger C, Waldmannstetter D, Lipkova J, Ezhov I, Tetteh G, et al. BraTS toolkit: translating BraTS brain tumor segmentation algorithms into clinical and scientific practice. Front Neurosci. 2020;14:125. https://doi.org/10.3389/fnins.2020.00125.
Langerak TR, van der Heide UA, Kotte AN, Viergever MA, van Vulpen M, Pluim JP. Label fusion in atlas-based segmentation using a selective and iterative method for performance level estimation (SIMPLE). IEEE Trans Med Imaging. 2010;29:2000–8. https://doi.org/10.1109/TMI.2010.2057442.
Galldiks N, Stoffels G, Filss C, Rapp M, Blau T, Tscherpel C, et al. The use of dynamic O-(2–18F-fluoroethyl)-l-tyrosine PET in the diagnosis of patients with progressive and recurrent glioma. Neuro Oncol. 2015;17:1293–300. https://doi.org/10.1093/neuonc/nov088.
Gottler J, Lukas M, Kluge A, Kaczmarz S, Gempt J, Ringel F, et al. Intra-lesional spatial correlation of static and dynamic FET-PET parameters with MRI-based cerebral blood volume in patients with untreated glioma. Eur J Nucl Med Mol Imaging. 2017;44:392–7. https://doi.org/10.1007/s00259-016-3585-0.
Breiman L. Random Forest Machine learning. 2001;45:5–32.
Sharma M, Juthani RG, Vogelbaum MA. Updated response assessment criteria for high-grade glioma: beyond the MacDonald criteria. Chin Clin Oncol. 2017;6:37. https://doi.org/10.21037/cco.2017.06.26.
Yang D. Standardized MRI assessment of high-grade glioma response: a review of the essential elements and pitfalls of the RANO criteria. Neurooncol Pract. 2016;3:59–67. https://doi.org/10.1093/nop/npv023.
Lutz K, Radbruch A, Wiestler B, Baumer P, Wick W, Bendszus M. Neuroradiological response criteria for high-grade gliomas. Clin Neuroradiol. 2011;21:199–205. https://doi.org/10.1007/s00062-011-0080-7.
Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20:728–40. https://doi.org/10.1016/S1470-2045(19)30098-1.
Sotoudeh H, Shafaat O, Bernstock JD, Brooks MD, Elsayed GA, Chen JA, et al. Artificial intelligence in the management of glioma: era of personalized medicine. Front Oncol. 2019;9:768. https://doi.org/10.3389/fonc.2019.00768.
Hutterer M, Hattingen E, Palm C, Proescholdt MA, Hau P. Current standards and new concepts in MRI and PET response assessment of antiangiogenic therapies in high-grade glioma patients. Neuro Oncol. 2015;17:784–800. https://doi.org/10.1093/neuonc/nou322.
Lohmeier J, Bohner G, Siebert E, Brenner W, Hamm B, Makowski MR. Quantitative biparametric analysis of hybrid (18)F-FET PET/MR-neuroimaging for differentiation between treatment response and recurrent glioma. Sci Rep. 2019;9:14603. https://doi.org/10.1038/s41598-019-50182-4.
Jena A, Taneja S, Gambhir A, Mishra AK, D’Souza MM, Verma SM, et al. Glioma recurrence versus radiation necrosis: single-session multiparametric approach using simultaneous O-(2–18F-fluoroethyl)-L-tyrosine PET/MRI. Clin Nucl Med. 2016;41:e228–36. https://doi.org/10.1097/RLU.0000000000001152.
Chang K, Beers AL, Bai HX, Brown JM, Ly KI, Li X, et al. Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro Oncol. 2019;21:1412–22. https://doi.org/10.1093/neuonc/noz106.
Ellingson BM. On the promise of artificial intelligence for standardizing radiographic response assessment in gliomas. Neuro Oncol. 2019;21:1346–7. https://doi.org/10.1093/neuonc/noz162.
Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. 2019.
Park KJ, Kim HS, Park JE, Shim WH, Kim SJ, Smith SA. Added value of amide proton transfer imaging to conventional and perfusion MR imaging for evaluating the treatment response of newly diagnosed glioblastoma. Eur Radiol. 2016;26:4390–403. https://doi.org/10.1007/s00330-016-4261-2.
Liesche F, Lukas M, Preibisch C, Shi K, Schlegel J, Meyer B, et al. (18)F-fluoroethyl-tyrosine uptake is correlated with amino acid transport and neovascularization in treatment-naive glioblastomas. Eur J Nucl Med Mol Imaging. 2019;46:2163–8. https://doi.org/10.1007/s00259-019-04407-3.
Schon S, Cabello J, Liesche-Starnecker F, Molina-Romero M, Eichinger P, Metz M, et al. Imaging glioma biology: spatial comparison of amino acid PET, amide proton transfer, and perfusion-weighted MRI in newly diagnosed gliomas. Eur J Nucl Med Mol Imaging. 2020;47:1468–75. https://doi.org/10.1007/s00259-019-04677-x.
Wiestler B, Kluge A, Lukas M, Gempt J, Ringel F, Schlegel J, et al. Multiparametric MRI-based differentiation of WHO grade II/III glioma and WHO grade IV glioblastoma. Sci Rep. 2016;6:35142. https://doi.org/10.1038/srep35142.
Pyka T, Hiob D, Preibisch C, Gempt J, Wiestler B, Schlegel J, et al. Diagnosis of glioma recurrence using multiparametric dynamic 18F-fluoroethyl-tyrosine PET-MRI. Eur J Radiol. 2018;103:32–7. https://doi.org/10.1016/j.ejrad.2018.04.003.
Hu X, Wong KK, Young GS, Guo L, Wong ST. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J Magn Reson Imaging. 2011;33:296–305. https://doi.org/10.1002/jmri.22432.
Funding
Open Access funding enabled and organized by Projekt DEAL. Wiestler B, Menze BH, and Kofler F are funded by SFB824.
Author information
Authors and Affiliations
Contributions
Conception and design: Wiestler B, Kirschke JS.
Acquiring data: Paprottka KJ, Kleiner S, Yakushev I.
Analyzing data: Paprottka KJ, Wiestler B, Preibisch C, Kofler F.
Drafting manuscript: Paprottka KJ, Wiestler B.
Revising the manuscript: Wiestler B, Kirschke JS, Paprottka KJ.
Approving the final content of the manuscript: All authors.
Corresponding author
Ethics declarations
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent for publication
All authors approved the article for publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paprottka, K.J., Kleiner, S., Preibisch, C. et al. Fully automated analysis combining [18F]-FET-PET and multiparametric MRI including DSC perfusion and APTw imaging: a promising tool for objective evaluation of glioma progression. Eur J Nucl Med Mol Imaging 48, 4445–4455 (2021). https://doi.org/10.1007/s00259-021-05427-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-021-05427-8