Abstract
The renewable, abundant , and low-cost nature of lignocellulosic biomass can play an important role in the sustainable production of bioenergy and several added-value bioproducts, thus providing alternative solutions to counteract the global energetic and industrial demands. The efficient conversion of lignocellulosic biomass greatly relies on the catalytic activity of carbohydrate-active enzymes (CAZymes). Finding novel and robust biocatalysts, capable of being active under harsh industrial conditions, is thus imperative to achieve an economically feasible process. In this study, thermophilic compost samples from three Portuguese companies were collected, and their metagenomic DNA was extracted and sequenced through shotgun sequencing. A novel multi-step bioinformatic pipeline was developed to find CAZymes and characterize the taxonomic and functional profiles of the microbial communities, using both reads and metagenome-assembled genomes (MAGs) as input. The samples’ microbiome was dominated by bacteria, where the classes Gammaproteobacteria, Alphaproteobacteria, and Balneolia stood out for their higher abundance, indicating that the degradation of compost biomass is mainly driven by bacterial enzymatic activity. Furthermore, the functional studies revealed that our samples are a rich reservoir of glycoside hydrolases (GH), particularly of GH5 and GH9 cellulases, and GH3 oligosaccharide-degrading enzymes. We further constructed metagenomic fosmid libraries with the compost DNA and demonstrated that a great number of clones exhibited β-glucosidase activity. The comparison of our samples with others from the literature showed that, independently of the composition and process conditions, composting is an excellent source of lignocellulose-degrading enzymes. To the best of our knowledge, this is the first comparative study on the CAZyme abundance and taxonomic/functional profiles of Portuguese compost samples.
Key points
• Sequence- and function-based metagenomics were used to find CAZymes in compost samples.
• Thermophilic composts proved to be rich in bacterial GH3, GH5, and GH9 enzymes.
• Compost-derived fosmid libraries are enriched in clones with β-glucosidase activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The environmental and human health concerns have compelled the replacement of fossil fuels with innovative and sustainable biobased supply chains ensuring economic viability and ecological compatibility. The biorefineries emerge as sustainable and green industrial processes aimed at the effective exploitation of residual biomass resources into bioenergy (biofuels, electricity, and heat) and other value-added products (biopolymers, biochemicals, biofertilizers, or bioplastics) (Duan et al. 2022). It is expected that the biobased product market will reach 50 billion euro by 2030 (Hassan et al. 2019).
Hence, lignocellulosic biomass, being a renewable, low cost, and highly abundant source of organic carbon, has the potential to become one of the world’s leading primary energy sources over the next century, also considering its annual global production (2 × 1011 t) (Devi et al. 2022). Lignocellulosic materials comprise food crops (e.g., sugarcane, corn, and soybean), non-food crops (e.g., eucalyptus, grasses, and willow), agroforestry residues (e.g., forest thinning, sugarcane bagasse, and wheat straw), agro-industrial wastes (spent coffee grounds, apple pomace, and municipal sludge), and inedible marine biomass (seaweeds and microalgae) (Hassan et al. 2019; Usmani et al. 2021). Although lignocellulosic residues differ between species, their main organizational components are cellulose (35 to 50%), hemicellulose (20 to 35%), and lignin (5 to 30%), which are arranged in a complex and rigid three-dimensional structure. The conversion of biomass into value-added products includes the following fundamental steps: (1) pretreatment to ease the access to the raw material (e.g., biological, chemical, and physical methods); (2) saccharification through enzymatic hydrolysis; (3) fermentation of sugars; and (4) purification of the end products (Ali et al. 2020). A set of diverse enzymes (carbohydrate-active enzymes, CAZymes) that are categorized into distinct classes and families are required to perform efficient lignocellulose depolymerization. These key enzymes can be classified into the glycoside hydrolases (GHs), auxiliary activities (AAs), glycosyltransferases (GTs), polysaccharide lyases (PLs), and carbohydrate esterases (CEs). Furthermore, useful carbohydrate-binding modules (CBMs) can also be associated to CAZymes (Cantarel et al. 2009; Braga et al. 2021).
There is an increasing need to find novel and robust enzymes with promising features that compete with those currently available on the market. The use of prominent techniques, which are independent of microbial cultivation, namely, metagenomics, allows to explore the genetic and metabolic diversity of complex ecosystems (Datta et al. 2020). Metagenomic studies are based on two main approaches: sequence-based and function-based metagenomics. The sequence-based approach is suitable to find new gene sequences that reveal similarities with the annotated genomes available in the databases. The functional approach allows the identification of unknown genes encoding novel bioactive molecules whose functions/activities would not be predicted only based on the DNA sequence (DeCastro et al. 2016; Madhavan et al. 2017; Datta et al. 2020).
Environmental DNA from soil, water, animal gut, industrial sewages, and extreme environments, such as composting, has been shown to have a great potential to uncover promising enzymes for biotechnological applications (Datta et al. 2020; Sousa et al. 2022). Due to the huge variety of microorganisms able to degrade lignocellulosic biomass involved in composting, it is considered one of the most important (bio)reactors that contribute to the renewable bioenergy on the planet. Some raw materials used in the composting process are green, agricultural, and agro-industrial residues, including animal feces, fruit, vegetable and crop harvesting residues, and municipal wastes. The whole composting process involves four phases, namely, the mesophilic phase, thermophilic phase, cooling phase, and maturation phase. The thermophilic phase, where temperatures above 65 °C can be reached, promotes the proliferation of thermophilic microorganisms able to produce interesting enzymes that degrade cellulose, hemicellulose, lignin, and proteins (Sánchez et al. 2017; Finore et al. 2023).
In this work, a sequence-based and functional metagenomic approach was implemented to identify, for the first time, carbohydrate-degrading enzymes in thermophilic compost samples containing lignocellulosic residues collected from three Portuguese composting units. To achieve this goal and also to obtain the taxonomic and functional profiles of the microbial communities, a multi-step bioinformatic pipeline herein developed was used. Additionally, the presence of β-glucosidase positive clones in the fosmid metagenomic libraries prepared with the DNA extracted from compost samples was experimentally validated. The clones herein described may thus provide excellent alternative biocatalysts that can be explored for a more efficient bioconversion of lignocellulosic biomass towards several industrial applications.
Materials and methods
Reagents
Cellulase from Trichoderma reesei, lipase from Candida rugosa, and all chemicals used in this work (analytical grade) were purchased from Sigma-Aldrich (St. Louis, USA). Proteinase K and RNase A were obtained from NZYTech (Lisbon, Portugal). Commercial humic acids and esculin were supplied by Alfa Aesar (Kandel, Germany). The CopyControl™ fosmid library kit, including the autoinduction solution, was provided by Epicentre Biotechnologies (Madison, Wisconsin, USA).
Sample collection and characterization
Compost samples with 1–2 kg were obtained in the thermophilic phase from three Portuguese composting companies, namely, Terra Fértil (TF), Lipor (LP), and Gintegral (GN) (Table 1). The sample from TF was collected in the unit of Parque Eco do Relvão, Chamusca (39° 24′ 11.4″ N 8° 23′ 11.9″ W). This composting unit mainly handles municipal sludge and agroforestry residues. The sample from LP was collected in Baguim do Monte, Porto (41° 11′ 58.4″ N 8° 32′ 46.6″ W). Lipor is the intermunicipal waste management service of Porto Metropolitan Area, and it works with food wastes from restaurants and local marketplaces and also green wastes from forests and public gardens. The sample from GN was obtained in the unit of Vitorino das Donas, Ponte de Lima (41° 43′ 17.3″ N 8° 39′ 35.3″ W). This composting unit mainly handles municipal sludge and several organic wastes considered unfit for direct application in agriculture. All the compost samples were stored at 4 °C until physicochemical characterization and DNA extraction.
The compost samples were characterized regarding their temperature, moisture, pH, elemental content, and humic acid concentration, as previously described by Costa et al. (2021). Temperature was determined on site with a thermometer. Briefly, to determine the moisture content, the compost samples were placed at 105 °C until reaching a constant weight. To measure the compost sample pH, a mixture of 4:1 (w/w) of water and compost was prepared, stirred for 30 min, and centrifuged to remove the non-soluble fraction, and the pH of the supernatant was then measured. The elemental analysis (total organic carbon, nitrogen, hydrogen, and sulfur) was performed by automated dry combustion (Requimte/LAQV, Faculty of Sciences and Technology, Nova University of Lisbon, Portugal). The concentration of humic acids was determined by absorbance measurement at 340 nm, using a standard curve (0–500 ng/μl) previously prepared with commercial humic acids, as described by Costa et al. (2021). Briefly, 1 g of compost samples was added to a 0.1 M NaOH solution (9 ml), stirred for 3 h at room temperature, and then centrifuged at 2500 × g for 10 min. The supernatant was acidified to pH 1.0 with HCl and incubated overnight in the dark (room temperature). The humic acids were then obtained by centrifugation (2500 × g, 10 min), air-dried, and resuspended in Tris–EDTA (TE) buffer. In all cases, the analyses were performed in triplicate. The data are presented as the means and respective standard deviations. GraphPad Prism (version 8.0.1.; https://www.graphpad.com/) was used to perform unpaired t-test. The same letters represent no significant differences for a 95% confidence level.
DNA extraction and metagenomic sequencing
The total metagenomic DNA was extracted from the compost samples using a methodology previously established by our group (Costa et al. 2021). This methodology is composed of three main steps, namely, the cell lysis and humic acid removal, the DNA recovery, and the DNA purification. Briefly, the compost samples (1 g) were firstly mixed with 5 ml of optimized lysis buffer containing 100 mM Tris-HCl, 100 mM Na EDTA, 1.5 M NaCl, 100 mM Na2HPO4, 100 mM CaCl2.2H2O, 1 mg/ml of proteinase K, 1 mg/ml of lysozyme, 0.2 mg/ml RNase A, 1% (w/v) powdered activated charcoal (4–8 mesh), and 1 g of glass beads (425–600 μm). After mixing using a vortex at maximum speed for 5 min, the samples were incubated for 30 min at 37 °C, 150 rpm. Then, 1 ml of 20% (w/v) sodium dodecyl sulfate was added, and samples were next incubated for 30 min at 65 °C. In a second step, the samples were centrifuged, and the supernatants were gently mixed with 1 volume of chloroform to isopropanol (C to I) (24:1 v/v). After centrifugation, the aqueous phase was used for DNA precipitation by the addition of 1 volume of C2H3NaO2 (3 M, pH 5.2) and 0.4 volumes of 30% (w/v) polyethylene glycol (MW-8000). DNA samples were initially kept at − 20 °C for 20 min and then slowly thawed on ice. The DNA pellet was precipitated by centrifugation and resuspended with 500 μl of TE buffer (10 mM Tris, 1 mM Na EDTA, pH 8.0). Finally, 1 volume of C to I (24:1 v/v) was added to the DNA and the mixture was centrifuged. The aqueous phase was transferred to a new tube followed by the addition of 1 volume of cold isopropanol. This mixture was then incubated for 5 min at 4 °C. The DNA was pelleted by centrifugation and washed twice with 500 μl of 70% (v/v) ethanol. The DNA samples were next centrifuged, and the pellet was air-dried for 10 min at room temperature. The metagenomic DNA samples were then dissolved in 100 μl of TE buffer and stored at 4 °C.
Metagenome analysis by shotgun sequencing was performed by Novogene (Cambridge, United Kingdom) using the Illumina NovaSeq6000 platform (Illumina, San Diego, CA, USA). The raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) database under the Bioproject PRJNA944686, with the following accession numbers: SAMN33758834 (TF sample), SAMN33758835 (GN sample), and SAMN33758836 (LP sample).
Metagenomic data analysis
The bioinformatic metagenomic pipeline used in this study is depicted in Fig. 1. Throughout the bioinformatic pipeline, different software and parameters were tested at each step, and those rendering more reliable and consistent output were selected to be used in the following steps of the pipeline. Raw sequencing data (raw reads) were checked for quality using FastQC software v. 0. 11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Clean reads were then used to perform taxonomic profiling with Kaiju v.1.9.0 (Menzel et al. 2016), using the nr_euk database (71 GB; 2022-03-10; link: https://kaiju.binf.ku.dk/server), which contains non-redundant proteins belonging to bacteria, archaea, and viruses, but also eukaryotic microorganisms and fungi, covering the most relevant taxonomic groups. Kaiju2table script was used to convert Kaiju’s output files into a summary table for taxonomic ranks.
Clean reads were then assembled into contigs using MEGAHIT v.1.2.9 (Li et al. 2015) and SPAdes v.3.15.4 (Bankevich et al. 2012). Following assembly quality assessment using QUAST software v.5.2.0 (Gurevich et al. 2013), where different parameters including assembly length, contig number, the largest contig, and N50 were evaluated, results from MEGAHIT were chosen for the subsequent analysis. Next, assembled contigs were grouped into bins, using three different binning tools: MaxBin2 (Wu et al. 2016), CONCOCT (Alneberg et al. 2014), and MetaBAT2 (Kang et al. 2019). Afterwards, Das Tool (Sieber et al. 2018) was employed to obtain the metagenome-assembled genomes (MAGs) from the combination of bins obtained by the three tools. For this purpose, only bins with a completeness above 50% and redundancy below 10% were considered. The taxonomic assignment of MAGs was also performed with Kaiju v.1.9.0 software.
Gene calling was performed in two rounds, considering MAGs obtained from Das Tool (Sieber et al. 2018) and also the clean reads. Two software were also used for gene calling—MetaGeneMark v. 3.25 (Zhu et al. 2010) and Diamond v.2.0.15 (Buchfink et al. 2015). A larger number of protein coding genes were obtained with Diamond, so these were used in the subsequent analysis.
Finally, functional annotation of protein coding genes was carried out using eggNOG v.2.1.6 (Jensen et al. 2008) and the CAZy database (Cantarel et al. 2009). Circos plots were created using Circos software v. 0.69.9 (Krzywinski et al. 2009).
Secondary data analysis
To perform a comparative analysis of our results with metagenomic data from diverse compost samples previously explored, different studies were chosen from the literature. This selection was based on the type of raw materials (mainly lignocellulosic residues), the composting process (homemade composting under uncontrolled conditions or handled composting with controlled environmental conditions), the time of sample collection (during the mesophilic or thermophilic phase of the composting process), and the performance of additional steps, such as enrichment with specific substrates of the compost samples. The exact same pipeline described in Fig. 1 was employed to analyze the reported data, with the exception of the omission of the binning step, since in this case, only the entire set of metagenomic contigs were used.
Construction of metagenomic fosmid libraries
The metagenomic DNA isolated from the three compost samples (TF, LP, and GN) was directly used in the preparation of three metagenomic libraries using the CopyControl™ fosmid library production kit (Biosearch Technologies, Hoddesdon, UK) and following the manufacturer’s instructions. Briefly, for each metagenomic library, the DNA fragment was end-repaired to generate blunt-ended, 5′-phosphorylated DNA and then, the size of the end-repaired DNA (approximately 40 kb) was confirmed by conventional agarose electrophoresis. Subsequently, the insert DNA was ligated with the linearized and dephosphorylated pCC1FOS™ vector, and the ligation product was packaged into lambda phage. Before plating the metagenomic libraries, the titer of the packaged phage particles was determined to obtain the desired number and density of clones in each plate (around 100 clones). Serial dilutions of packaged phage particles were prepared in phage dilution buffer (PDB) (10 mM Tris-HCl pH 8.3, 100 mM NaCl, 10 mM MgCl2) and infected phage T1–resistant EPI300 T1R Escherichia coli cells (trfA gene, required for replication initiation, was engineered to be under tight control of an inducible promoter) were spread on lysogeny broth (LB) agar plates supplemented with 12.5 μg/ml chloramphenicol and grown overnight at 37 °C. Five hundred sixty-three clones from each metagenomic library were isolated and grown in LB medium supplemented with 10 mM MgSO4, 2 g/l maltose, 12.5 μg/ml chloramphenicol, and 0.2% (v/v) CopyControl™ fosmid autoinduction solution (500 ×). Each metagenomic library was stored at − 80 °C in supplemented LB medium with 20% of glycerol.
β-Glucosidase activity screening and comparison with CAZy prediction
The functional screening of the three metagenomic libraries to detect the β-glucosidase activity was based on a practical and rapid assay performed on 96-well microplates containing a suitable substrate (esculin) for this enzymatic activity.
After the growth of the 563 clones in LB medium supplemented with 12.5 μg/ml chloramphenicol, 10 mM MgSO4, 2 g/l maltose, and 0.2% (v/v) CopyControl™ fosmid autoinduction solution (500 ×), they were transferred to 96-well microplates with LB agar, 12.5 μg/ml chloramphenicol, 10 g/l arabinose, 0.1 g/l esculin, and 0.5 g/l ferric chloride. After incubation at 37 °C for 24 h, the 96-well microplates were kept at room temperature (approximately, 25 °C) for 1 week. A color change of the agar culture medium to brown was considered a positive response. Commercial enzymes, namely, cellulase from T. reesei and lipase from C. rugosa, were used as positive and negative controls, respectively.
In order to compare the results predicted through the CAZy database with the data obtained in the functional screening for β-glucosidase activity, the number of positive clones obtained in the experimental screening after 1 week was compared with the number of predicted CAZymes belonging to the GH1, GH3, GH5, GH9, and GH30 families, which are known to contain β-glucosidases (Cairns and Esen 2010).
Results
Physicochemical characterization of the composting samples
Table 1 presents information about the three compost samples collected from TF, LP,, and GN, together with the results obtained for their physicochemical characterization.
As shown in Table 1, three compost samples were obtained from different composting units, which used mixtures of distinct types of input wastes in different proportions. In general, all samples were composed of agroforestry residues or derive from them, and the compost samples from TF and GN contained a significant percentage of municipal sludges. Additionally, the samples from LP and GN presented similar sampling temperatures, while for the TF compost, a higher temperature was recorded (62.7 ± 1.0 °C). Nevertheless, all samples were collected in the thermophilic phase (temperature > 45 °C) of the composting process. Regarding the moisture content, the LP sample presented the lowest value (59.55 ± 1.93%), although not statistically different from the GN sample (63.44 ± 2.20%). In contrast to this parameter, significant statistical differences were obtained for the pH, being the highest value determined for the LP sample (8.92 ± 0.08). For the elemental analysis, significant statistical differences were only found for the TF and LP samples. In all samples, a higher percentage of total carbon (between 13 and 20%) and a lower fraction of total sulfur (< 0.5%) were observed. The total nitrogen values were in all cases in the range of 1–2%. The total hydrogen percentage was lower for the TF compost (4.22 ± 0.21%) and higher for the GN compost (9.56 ± 1.80%). A C to N ratio of approximately 11 was observed for the three samples. Finally, the humic acid content obtained from these samples was statistically different, and the highest value (18.42 ± 0.11 mg∙g−1 compost) was determined for the LP sample.
General features of the metagenome
Metagenome sequencing on the Illumina NovaSeq6000 platform yielded 89.8, 79.9, and 96.3 million raw reads for the TF, LP, and GN samples, respectively, with 150 bp each (Table 2). After the de novo assembly using MEGAHIT, the TF metagenome was found to be composed of 349,015 contigs (186,612 of them with more than 500 bp and the longest having 702,260 bp), with an average GC content of 55.6%, a N50 value of 3990 bp, and with 464 scaffolds higher than 5000 bp. The LP metagenome has 1,055,919 contigs (536,996 of them with more than 500 bp and the longest with 916,338 bp), average GC content of 55.9%, N50 of 1919 bp, and 425 scaffolds higher than 5000 bp. As for the GN metagenome, it has 1,027,319 contigs (530,791 of them with more than 500 bp, contigs, having the longest one 1833 bp), an average GC content of 56.8%, a N50 of 1991 bp, and 663 scaffolds higher than 5000 bp. About 40, 34, and 35% of the assembled contigs can be recruited to the contigs greater than 1000 bp for the TF, LP, and GN samples, respectively. Gene calling based on the assembled contigs using DIAMOND algorithm predicted 677,723 protein coding sequences (CDSs) for the TF sample and more than 1.7 million CDSs for the LP and GN samples, with an average length of 188.2, 168.7, and 170.5 amino acids, respectively. A total of 60.2 and 82.7% of the TF contigs were annotated by the COG (Tatusov et al. 2000) and KEGG (Kanehisa and Goto 2000) databases, respectively. For the LP and GN samples, these percentages were 62.1 and 83.1% (LP) and 64.4 and 61.6% (GN), respectively. Importantly, in the assembled metagenomic contigs of the three samples, 0.91–1.1% were annotated as CAZymes (CAZy database (Cantarel et al. 2009)).
Taxonomic composition of compost microbiota
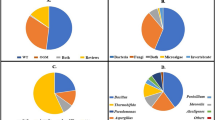
The taxonomic classification of the metagenomic samples was performed with the clean reads (prior to assembly) using the Kaiju software (Menzel et al. 2016) and the nr_euk database. For the three samples, around 70% of the reads could be assigned to a taxon, leaving only around 30% that could not be assigned to any community, which validates the use of the software and of the database. Results from domain analysis (Fig. 2A) showed that all samples were clearly dominated by Bacteria (98.3–98.9%). In the TF sample, this percentage was followed by Viruses (0.9%), Archaea (0.1%), and Eukaryota (0.07%). As for the LP sample, Bacteria higher abundance was followed by Viruses (1%), Eukaryota (0.6%), and Archaea (0.07%). Finally, in the GN sample, Eukaryota (1.4%) followed Bacteria in terms of relative abundance and then Viruses (0.3%) and Archaea (0.05%). The taxonomic analysis also revealed a similar number of species, genera, families, orders, and classes among the three samples, with mean values of 32,740 species; 4189 genera; 1019 families; 473 orders; and 200 classes (Supplemental Tables S1–S6).
In terms of class taxonomic assessment (Fig. 2B), the TF sample was dominated by Gammaproteobacteria (31.6%) and Balneolia (21.6%), with these two classes accounting for more than 50% of the community and being this high amount of Balneolia bacteria exclusive of this sample. Flavobacteriia (16.6%) and Alphaproteobacteria (11.3%) were the next two most representative classes in this sample. The predominant class in the LP sample was Alphaproteobacteria (30.9%), which was also only observed for this sample, followed by Gammaproteobacteria (22.8%) and Flavobacteriia (14%). Regarding the GN sample, the class Gammaproteobacteria (31.4%) exhibited the highest relative abundance, as it was also detected in the TF sample, followed by Alphaproteobacteria (19%), Betaproteobacteria (12.9%) and Sphingobacteriia (12.4%). Regarding the order of taxonomic identification, a very heterogenous pattern was observed between samples. The most abundant orders (Fig. 2C) were Balneolales (23.1%) in the TF sample, Rhizobiales (16.2%) and Flavobacteriales (15.7%) in the LP sample, and Xanthomonadales (21.5%) in the GN sample.
Functional profiles and metabolic pathway annotation of the metagenomes
To analyze the metagenomes’ functional profiles, after assembly, the putative protein coding genes identified using DIAMOND algorithm were mapped against the COG database (Tatusov et al. 2000). In all three samples, the most predominant functional category was “metabolism” followed by an identical percentage of sequences belonging to the categories “cellular processes and signalling” and “information storage and processing” (Fig. 3). Only around 20% of the sequences were assigned to “unknown function” in all samples (Fig. 3A). Within the “metabolism” category, the most abundant COG categories for all samples were “amino acid transport and metabolism,” followed by “energy production and conversion,” “inorganic ion transport and metabolism,” “carbohydrate transport and metabolism,” and “lipid transport and metabolism.” Within the “cellular processes and signalling” and “information storage and processing” categories, the most abundant COG categories were “cell wall/membrane/envelope biosynthesis” and “replication, recombination and repair,” respectively (Fig. 3B). In terms of sample comparison, no large differences were detected between samples.
Functional and metabolic classification based on COG and KEGG databases for Terra Fértil (TF), Lipor (LP) and Gintegral (GN) samples. (A) Distribution of the main categories of the COG database across the three samples. (B) Distribution of COG sub-categories across the three samples. (C) Distribution of genes among level 1 KEGG metabolic pathways for the TF, LP and GN samples. Distribution of genes among level 2 KEGG metabolic pathways for the TF (D), LP (E) and GN (F) samples.
Annotation of the metagenomic contigs against the KEGG database (Kanehisa and Goto 2000) was performed to identify the putative genes mapped to different metabolic pathways. In agreement with the annotation against the COG database, pathways related to “metabolism” were by far the most abundant among the three samples, with the LP and GN samples displaying a much higher gene count within this category than the TF sample (Fig. 3C). The GN sample showed the highest number of genes associated with “metabolism,” being this number more than three times higher than the one found in the TF sample. Additionally, the “carbohydrate metabolism”–related pathways were those gathering a higher number of putative genes in all samples, followed by “amino acid metabolism,” “energy metabolism,” “metabolism of cofactors and vitamins,” and “xenobiotics biodegradation and metabolism.” Within the other level 2 KEGG pathways, a high number of genes were mapped to the pathways associated with “membrane transport” and “signal transduction” for all samples and “neurodegenerative diseases” for the GN and LP samples (Fig. 3D).
The combined analysis of functional annotation against KEGG and COG databases revealed a clear dominance of the contigs associated with metabolism on the compost metagenomic samples, with particular emphasis on carbohydrate metabolism.
Mining of carbohydrate-active enzymes
In order to identify putative enzymes able to degrade lignocellulosic biomass in the metagenomes, the reads were queried against the CAZy database (Cantarel et al. 2009; Drula et al. 2022), which is dedicated to enzymes involved in the building and breakdown of complex carbohydrates (CAZymes). Among the protein coding genes, around 6170; 19,000; and 16,470 for the TF, LP, and GN samples, respectively, were mapped to this database corresponding to approximately 1% of all the predicted genes (Table 2).
The CAZy classification comprises 5 classes of enzymatic activities: GHs that include glycosidases and transglycosidases; GTs which can be either retaining or inverting enzymes; PLs that include enzymes with an ability to cleave uronic acid–containing polysaccharide chains; CEs which catalyze the de-O or de-N-acylation of substituted saccharides; and AAs that describe a wide variety of enzyme mechanisms and substrates associated with lignocellulose conversion including lignin degradation. Additionally, carbohydrate-active enzymes often display non-catalytic modules, the CBMs, which are protein fragments that lack enzymatic activity per se but potentiate the activity of the aforementioned enzymes (Lombard et al. 2014). The 6 enzymatic activities were detected in all samples revealing the potential of the putative genes encoded in the compost metagenomes for lignocellulose degradation. Considering the total number of genes from the three samples assigned to each individual family, it is evident that the LP and GN samples displayed a higher number of CAZymes when compared with the TF sample (Fig. 4A).
Distribution of carbohydrate-active enzymes (CAZymes) in the Terra Fértil (TF), Lipor (LP), and Gintegral (GN) samples. A Contribution of each sample to the total number of protein coding sequences assigned to each CAZyme family. B Distribution of CAZymes in the three samples. The inner ring describes the main CAZyme classes (GT, green; GH, dark blue; CBMs, orange; CE, purple; AA, light blue; PL, light yellow), while the outer ring depicts the families, as well as the corresponding number of protein coding sequences annotated. C Circos plot depicting the relative abundance of CAZymes in the TF, LP, and GN samples. The inner ring indicates the total number of protein coding sequences associated to each sample or CAZyme class; the outer ring represents the relative abundance of protein coding sequences from each sample or CAZyme class; the width of the bars connecting a given sample and CAZyme class indicates their relative abundance to each other. GH glycoside hydrolases, GT glycosyltransferases, PL polysaccharide lyases, CE carbohydrate esterases, CBMs carbohydrate-binding modules, AA auxiliary activities
Considering the relative abundance of each enzymatic family in the metagenomic samples, clearly, the three samples were dominated by GHs and GTs (Fig. 4B and C). The GT2 family was the most abundant in all composting samples, with GT51 and GT4 also counting with many putative genes from the metagenomes (Fig. 4B). For GHs, the GH13 family was by far the most abundant in all samples, followed by GH3, GH5, GH9, and GH31 (Fig. 4B).
Regarding the taxonomic classification of the putative enzymes found in the metagenomes, CAZymes largely belonged to the Bacteria domain (Fig. 5A), being particularly distributed through 17–22 bacterial phyla (Fig. 5B and C). Importantly, in all samples, the four most abundant CAZyme classes (GHs, GTs, CBMs, and CEs) were clearly affiliated to three different bacterial phyla: Proteobacteria, Bacteroidetes, and Actinobacteria (Fig. 5B and C). Though the great majority of CAZymes found in our samples were bacterial, among the domains Archaea and Eukarya, the phylum Euryarchaeota and the kingdom Fungi showed higher abundances in both GHs and GTs, respectively, except for the TF sample that had a higher portion of GHs in the Metazoa kingdom (Supplemental Fig. S1).
Taxonomic distribution of bacterial CAZymes in the Terra Fértil (TF), Lipor (LP), and Gintegral (GN) samples. A Distribution of protein coding sequences (CDSs) annotated to the CAZy database among the three domains of life (Bacteria, Archaea, and Eukarya) in the three compost samples. B Circos plots depicting the taxonomic affiliation of CAZymes in the TF, LP, and GN samples. The enzyme classes are on the right side and the phyla numbers on the left; the inner ring represents the total number of protein coding sequences associated to each CAZyme class or phylum; the outer ring represents the relative abundance of protein coding sequences from each CAZyme class or phylum; the width of the bars connecting a given CAZyme class and phylum indicates their relative abundance with respect to each other. C Support table that identifies the phylum each number corresponds in the circos plots depicted in B for each sample. GH glycoside hydrolases, GT glycosyltransferases, PL polysaccharide lyases, CE carbohydrate esterases, CBMs carbohydrate-binding modules, AA auxiliary activities
Lignocellulose degradation potential of MAGs retrieved from compost samples
To identify the microorganisms with higher potential for lignocellulose degradation within the collected compost samples, binned MAGs with completeness ≥ 50% and redundancy < 10% were recovered and further analyzed regarding taxonomy, functionality, and presence of CAZymes. A total of 44, 66, and 69 of MAGs that fulfilled these parameters were recovered from the TF, LP, and GN samples, respectively. The features and statistics of MAG assemblies, as well as the maximum predicted taxonomic level are summarized in Supplemental Table S7. The average size of MAGs was around 3.2 Mb with a mean N50 of 38,900 bp. The GC content varied from 33.2 to 74.4%, indicating a broad range of microorganisms.
Figure 6A and B shows the taxonomic inference of MAGs at the class and order levels. It is worth mentioning that not all MAGs could be identified at these levels (Supplemental Table S7). Indeed, 40.9–45.5% of MAGs could not be classified at the class level and 60.6–63.6% at the order level. Nevertheless, all the putative genomes were assigned to the Bacteria domain (Supplemental Table S7) (Fig. 2A). Taking into account the MAGs classified at the class level, the TF sample was clearly dominated by bacteria belonging to the Gammaproteobacteria class, followed by Alphaproteobacteria, Flavobacteriia, and Balneolia. The LP and GN samples had higher percentage of bacteria belonging to Alphaproteobacteria, followed by Gammaproteobacteria. However, while the third most abundant class in the LP sample was Flavobacteriia, in the GN sample, it was Sphingobacteriia (Fig. 6A). At the order level, the two most abundant in the TF sample were Flavobacteriales and Balneolales accounting for nearly 20% of the orders represented in the repertoire of MAGs from this sample. As for the LP sample, it was dominated by Flavobacteriales (25.9%) followed by Cytophagales (14.8%). Lastly, the GN sample had a higher abundance of Sphingobacteriales (20%) followed by Rhizobiales (16.7%) (Fig. 6B).
Taxonomic classification, functional annotation, and identification of CAZymes in the metagenome-assembled genomes (MAGs) recovered from Terra Fértil (TF), Lipor (LP), and Gintegral (GN) samples. A Taxonomic distribution of MAGs that could be assigned to the class level. Not classified MAGs were excluded from the calculation of abundancy. B Taxonomic classification of MAGs that could be affiliated at the order level. Not classified MAGs were excluded from the calculation of abundancy. C Functional annotation of MAGs recovered from the three samples based on the main categories of the COG database. D Distribution of COG sub-categories across the MAGs recovered from three samples. E Circos plot depicting the relative abundance of CAZymes in MAGs recovered from the TF, LP, and GN samples. The inner ring indicates the total number of protein coding sequences associated to each sample or CAZyme class; the outer ring represents the relative abundance of protein coding sequences from each sample or CAZyme class; the width of the bars connecting a given sample and CAZyme class indicates their relative abundance to each other. GH glycoside hydrolases, GT glycosyltransferases, PL polysaccharide lyases, CE carbohydrate esterases, CBMs carbohydrate-binding modules, AA auxiliary activities. F Presence of the GH families associated with lignocellulose-degrading enzymes in the MAGs recovered from the three samples. The heatmap represents the number of protein coding sequences from each MAG mapped to each family. Only MAGs displaying at least five protein coding sequences belonging to any GH family were included. The taxonomic identification at the phylum level is also depicted, except for the LP43 to which the maximum taxonomic level obtained was the kingdom
Of note, 9 MAGs could be assigned at the species level. Specifically, in the TF sample, 2 MAGs were affiliated to Lysobacter defluvii, 1 MAG to Pedobacter indicus, and 1 MAG to Galbibacter marinus. In the LP sample, 2 MAGs were assigned to Bradymonas sediminis, 1 MAG to Paracoccus alcaliphilus, and another one to Rhodothermus marinus. In the GN sample, 1 MAG was identified as Pseudofulvimonas gallinarii (Supplemental Table S7).
Functionally, the annotation using eggNOG revealed that MAGs collected from the three samples were very similar with the most represented COG category being “metabolism” (Fig. 6C) and with the most abundant sub-categories being “amino acid transport and metabolism,” followed by “energy production and conversion,” “inorganic ion transport and metabolism” and “carbohydrate transport and metabolism” for all samples (Fig. 6D). These data indicate that the differences observed at physicochemical characterization did not affect their overall functional behavior.
To unveil the lignocellulose degradation potential of the MAGs recovered from the compost samples, MAGs were queried against the CAZy database to identify the genes encoding lignocellulolytic enzymes. Results were in agreement with those obtained with clean reads as they showed that, in all samples, the most represented CAZyme families were GHs and GTs (Fig. 6E). Indeed, an average of 25.4 and 16.6 CDSs affiliated to GHs and GTs, respectively, were identified in the compost samples’ MAGs (Supplemental Table S7).
Among CAZymes, GHs are of particular interest as they include a wide range of enzymes useful for the utilization of lignocellulosic biomass (Gong et al. 2020; Reichart et al. 2021). Therefore, the abundance of GH families related to cellulases, endo-hemicellulases, debranching enzymes, and oligosaccharide-degrading enzymes in the retrieved MAGs was studied in detail (Fig. 6F, Supplemental Table S7). Figure 6F shows the MAGs that had at least five CDSs affiliated to any GH family related to the aforementioned categories. It is noteworthy that the LP and GN samples were enriched in MAGs that fulfill these criteria (7–8) in comparison with the TF sample, which was only represented by 3 MAGs. Overall, the genes encoding lignocellulolytic enzymes were distributed through the following families: cellulases (GH5, GH6, GH9), endo-hemicellulases (GH26), oligosaccharide-degrading enzymes (GH3, GH29, GH43), and debranching enzymes (GH51). Among these families, GH5 and GH9 clearly stood out as all the depicted MAGs displayed at least 5 CDSs belonging to these families, except for the LP16, and with particular abundance in the LP43 and GN22 MAGs (Fig. 6F).
Considering the taxonomic affiliation, 50% of the MAGs enriched in lignocellulose-degrading enzymes belonged to the phylum Bacteroidetes, followed by Proteobacteria (22.2%). Additionally, 2 MAGs assigned to the phylum Actinobacteria, 1 to Balneolaeota, and 1 to Deinococcus-Thermus also showed an interesting abundance of these enzymes. Notably, the Balneolaeota TF23 MAG, a phylum that was only present in the TF sample, exhibited GHs belonging to all the main categories represented in the samples from this study. Curiously, there was no clear distribution of a particular GH family across a particular phylum, but some families appeared only in a single MAG. Indeed, the α-mannosidases GH38 were only present in the GN53 MAG (Bacteroidetes), the β-xylosidases GH39 in the GN19 MAG (Actinobacteria), and the cellobiose phosphorylases GH94 in the LP43 MAG (could only be assigned to the Bacteria kingdom). In addition, the endo-1,4-β-xylanases GH8 appeared only in the LP16 (Proteobacteria) and GN22 (Actinobacteria) MAGs (Fig. 6F).
Diversity of lignocellulose-degrading glycoside hydrolases in compost metagenomes from diverse origins
To compare the potential for lignocellulosic biomass conversion of the samples herein described with other compost samples holding different features and composition, we next analyzed the abundance of the GH families more important for plant biomass processing (Gong et al. 2020; Reichart et al. 2021) in the TF, LP, and GN samples and also in composting samples selected from the literature. For that purpose, the metagenomic sequencing data from the selected samples were processed using the same pipeline applied in this work (Fig. 1, Supplemental Table S8) and a particular set of genes encoding for four categories of lignocellulose-degrading enzymes, namely, cellulases, endo-hemicellulases, debranching enzymes, and oligosaccharide-degrading enzymes (Table 3), was evaluated, as described by Gong et al. (2020) and Reichart et al. (2021).
A detailed description of the selected samples can be found in Supplemental Table S9. Sample SRR032208 (SG) was collected from switchgrass feedstock inoculated in bioreactor with green-waste compost (Allgaier et al. 2010); sample SRR10856164 (CBLU) was harvested from a pilot experiment performed in ventilated compartments of compost containing corncob, bran, lime, and urea (Kong et al. 2020); samples SRR4381490 (AD), SRR4381491 (EC) and SRR4381492 (PN) were collected from piles of chipped wood from Arundo donax, Eucalyptus camaldulensis, and Populus nigra, respectively, subjected to natural biodegradation under oak trees (Montella et al. 2017); and finally, samples SRR2388864 (FN1), SRR2388865 (FN2) and SRR2388866 (FN3) were harvested after 2, 7, and 19 days of cultivation, respectively, of a symbiotic cellulolytic microbial consortium based on forest compost soil that was subjected to successive enrichment on Napier grass under facultative anoxic conditions (Kanokratana et al. 2018).
The LP and EC samples were those with higher number of CDSs annotated as GHs (> 10,000) followed by the GN and AD samples (> 8000). Analysis of the abundance of GHs in relation to the total number of CDSs annotated to the CAZy database revealed that all compost samples have a similar percentage of GHs ranging from 49.1% in the CBLU sample to 62.4% in the FN3 sample (Table 3), attesting the widely recognized richness of lignocellulolytic enzymes present in compost samples (Allgaier et al. 2010; Montella et al. 2017; Kanokratana et al. 2018; Kong et al. 2020). The most abundant category in the TF and LP samples was cellulases (15.2 and 16.7%, respectively), while the GN sample from this study and all the samples selected from the literature mainly contained oligosaccharide-degrading enzymes (17.5–31.2%). On the other hand, as above described for MAGs, the most prevalent families of GHs (5.7–8.2%) in our samples were the cellulases GH5, the endoglucanases GH9, and the β-glucosidases GH3. Interestingly, the same families were also among the most represented in the samples selected for comparison (4.9–16.3%). However, in addition to those, the CBLU, PN, and FN1 samples were further enriched in the β-xylosidases belonging to the family GH43 (5.1–5.6%) and the FN2 and FN3 samples in the α-L-fucosidases of the GH29 family (5.5–6.2%). The SG and CBLU samples were also abundant in debranching enzymes with α-L-arabinofuranosidase activity from the GH51 family (5.4–6.0%). Though much less represented, genes encoding for endo-hemicellulases of the GH8, GH26, and GH28 families with endo-1,4-β-xylanase, xyloglucanase, and polygalacturonase activities, respectively, were also found in the selected compost samples (Table 3).
β-Glucosidase activity in the metagenomes of compost samples
Since GHs are one of the main CAZyme families represented in the compost metagenomes and given that they hold a great potential for lignocellulose biomass degradation, we next decided to construct metagenomic fosmid libraries with the DNA extracted from the three compost samples (TF, LP, and GN), to experimentally validate the presence and abundance of these enzymes in the collected samples. Since β-glucosidase (which belongs to the GH family) activity is often a limiting factor in cellulase conversion and glucose release (Zang et al. 2018), the three compost-derived metagenomic libraries were analyzed through a functional screening performed on agar microplates containing esculin, the specific substrate for the β-glucosidase activity.
In this way, 563 clones from each metagenomic library were evaluated over 1 week and the observation of brown color was considered a positive response. All libraries exhibited a good number of positive clones for β-glucosidase activity, attesting the presence of these enzymes in the samples, in agreement with the sequencing approach. The metagenomic library constructed from the GN sample (Fig. 7C) presented a higher number of clones with β-glucosidase activity (79), followed by the LP library (71) and finally, the TF library with 59 positive clones (Fig. 7A and B). To compare the experimental data with the annotation against the CAZy database, the percentage of positive clones in each library was compared with the number of CDSs in the GH1, GH3, GH5, GH9, and GH30 families, which include β-glucosidases (Cairns and Esen 2010). Results show that compost metagenomes display around 19–22% of CAZymes belonging to the specified GH families, while 9–14% of clones were positive for β-glucosidase activity (Fig. 7D), effectively demonstrating the presence of these enzymes in the compost samples.
Experimental validation of β-glucosidase activity in the compost metagenomic libraries in comparison with β-glucosidase families predicted using the CAZy database. Visual representation of clones with positive response to the β-glucosidase activity for the three metagenomic libraries from A Terra Fértil (TF), B Lipor (LP), and C Gintegral (GN) compost samples. D Relative abundance of clones with β-glucosidase activity in the experimental screening and predicted CAZymes belonging to the GH1, GH3, GH5, GH9, and GH30 families in the TF, LP, and GN metagenomes
Discussion
Exploring the aerobic composting ecosystem has been proved to be an interesting approach to find novel microorganisms and identify promising metabolites, namely, enzymes stable and active under harsh conditions. The composting samples studied in this work (TF, LP, and GN) presented high temperatures, since they were all collected specifically in the thermophilic phase of the composting process. The changes which occur at this phase are dependent on the growth and metabolism of thermophilic microorganisms, being affected by certain parameters, such as moisture, total carbon, and nitrogen contents. The significant differences obtained for these particular parameters between the TF and LP samples may be related with the presence of municipal sludges in the TF sample. Municipal sludges are rich in nitrogen and are characterized by high moisture content and low porosity (Kulikowska et al. 2022). Both, the TF and GN samples, presented a high percentage of municipal sludges and moisture content (> 63%). On the other hand, the moisture content (around 60%) recorded in the LP sample may be mainly associated with the inclusion of green and food residues which, in addition to the moisture, have also a high nitrogen content (Palaniveloo et al. 2020). The mixture of green organic wastes with lignocellulosic residues (brown residues) may have contributed to the differences observed in the total carbon and nitrogen contents. Green wastes generally present higher nitrogen content, while brown residues are mainly rich in carbon (Hemati et al. 2022; Kulikowska et al. 2022). For the three samples, the C to N ratios did not differ significantly, being obtained values around 11. Nevertheless, the samples studied in this work presented lower C to N ratios than those generally reported in the literature (Goyal et al. 2005; Awasthi et al. 2014; Chin et al. 2020). The lower C to N ratios together with the high moisture contents may explain the longer periods, due to a slower degradation, to reach the typical temperatures of the thermophilic phase of the composting process. Usually, temperatures above 50 °C are reached after a week (Guo et al. 2012; Awasthi et al. 2014; Zhang et al. 2020; Kulikowska et al. 2022). Regarding the pH variations, the TF and GN samples presented values close to 7 that benefit the microbial activities responsible for the decomposition of organic matter to produce organic acids and inorganic salts. The alkaline pH measured in the LP sample can be related to the type of residues (food wastes) that composed the sample, since an increase in pH is generally due to ammonia release as a consequence of protein degradation (Palaniveloo et al. 2020; Zhang et al. 2020). The humic acid content was significantly different for the three samples. The higher humic acid content in the LP sample can be related to a higher organic carbon content released from food wastes as they are more easily degraded than other recalcitrant materials. Also, the later sampling time of the LP sample (4 weeks) compared to the TF and GN samples can explain the amount of humic acids obtained, since these acids are generated over time according to the degradation of simpler components, followed by cellulose, hemicellulose, and lignin. On the other hand, the lower humic acid content in the GN sample may be associated with both the shorter sampling period and the sample composition. This sample was collected only after 2–3 weeks of the composting process, and it is known that humic acids can be used as substrate by thermophilic microorganisms in the early stages of the thermophilic phase (Unsal and Sozudogru Ok 2001; Hemati et al. 2022; Zhang et al. 2022), thus not being accumulated in the medium. Additionally, the GN sample presented a lower percentage of lignocellulosic materials and included cork chips in its composition, which is generally described as a very recalcitrant material for microbial degradation (Martins et al. 2014). All these facts might have affected the humic acid concentration obtained for the GN sample.
According to the literature, the process of organic matter degradation is essentially carried out by thermophilic microorganisms represented mainly by bacteria and fungi (Palaniveloo et al. 2020). In fact, our samples were clearly dominated by bacteria. The greater abundance of Alphaproteobacteria and Gammaproteobacteria (phylum Proteobacteria) was common for the three samples. The bacterial strains belonging to the phylum Proteobacteria are known to adapt to harsh ecosystems, namely, composting at the thermophilic phase (Zhou et al. 2017; Kaushal et al. 2018). The mentioned classes specifically harbor a wide variety of genes responsible for enhancing the degradation process of lignocellulose, as well as residual sludges, being the type of microorganisms commonly associated to the carbon and nitrogen cycles. In addition, they can break small molecules, such as glucose, propionate, and butyrate, and are positively related to neutral pH. All this evidence is in agreement with the characteristics of our samples. The low abundance of Betaproteobacteria in the TF and LP samples may explain the slower decomposition rate to reach high temperatures compared to the GN sample (Ma et al. 2020; Wang et al. 2020; Cao et al. 2021). Additionally, the two classes Sphingobacteria and Flavobacteriia (phylum Bacteroidetes) are also recognized for exhibiting a crucial role in the breakdown of recalcitrant polysaccharides in lignocellulose biomass (Le et al. 2022). The greater taxonomic difference was found in the TF sample, which presents in its composition an additional class, Balneolia, belonging to the phylum Balneolaeota (Costa et al. 2021). The members of this phylum are extremophiles capable of surviving and play an important role in extreme salt, temperature, or pH environments (Hahnke et al. 2016; Jo et al. 2021). The abundance of this class may be related to the higher temperature (62.7 °C) of the TF sample when compared to the LP and GN samples (50–52 °C), resulting in suitable conditions for the activity of this type of thermophilic microorganisms. However, other studies have reported the growth of microorganisms belonging to the Balneolia class in saline environments, namely, salterns and wastewater with high salinity (Jo et al. 2021; Almeida et al. 2022). This evidence may suggest that the residues used in the composting process of the TF sample, in particular municipal sludge, could present a salinity content that allows the development of this type of microorganisms.
Looking to the repertoire of MAGs, the taxonomic classification was very similar as all of them were classified into the Bacteria domain, confirming the results obtained before assembly. The abundance of Gammaproteobacteria and Alphaproteobacteria was also a common feature, as it was the presence of MAGs belonging to Flavobacteriia, Sphingobacteriia, and Balneolia, the latter only present in the TF sample. The large discrepancy between Bacteria and the other domains in terms of abundance and diversity of taxonomic units, both in the analysis of clean reads and MAGs, may arise not only from the sample characteristics but also from the chosen taxonomic database. Even though the kaiju nr_euk database was chosen as an attempt to characterize other microorganisms’ domains, most metagenomic databases include mainly bacterial data, or at least the most well-characterized non-bacterial taxa, so within the 30% of the unclassified reads, we need to account for the possible existence of less known and even novel non-bacterial taxa. Other databases and software were employed to expand the taxonomic characterization of the metagenomes, although with scarce results. Interestingly, the number of genomes that could be taxonomically classified at the species level was significantly decreased in all samples, suggesting that most MAGs herein reconstructed belong to undiscovered species, which require future endeavors to prompt their taxonomic classification. However, 7 species were assigned to the 9 MAGs and most of them were previously isolated from extreme environments, such as geothermal resources, R. marinus (Alfredson et al. 1988); deep-water/coastal areas, P. indicus (He et al. 2020), G. marinus (Li et al. 2013), and B. sediminis (Wang et al. 2015); and municipal solid waste landfills, L. defluvii (Yassin et al. 2007). In addition, the species L. defluvii (Hayat et al. 2013), G. marinus (Ballardo et al. 2020), P. alcaliphilus (Tian et al. 2013), R. marinus (Braga et al. 2021), and P. gallinarii (Song et al. 2015) are already related to biomass-degradation processes, namely, composting. These species have been recognized as crucial lignocellulose degraders given their ability to produce polysaccharide-degrading enzymes (e.g., cellulases, xylanases, and amylases) capable of acting under severe conditions, such as temperature and pH (Gomes et al. 2003; Hayat et al. 2013; Ballardo et al. 2020). These facts agree with the sampling temperature and pH of our samples. For instance, P. alcaliphilus, alkaliphilic bacteria (Urakami et al. 1989), was specifically found in the LP sample that differs from the TF and GN samples by its alkaline pH.
Consistent with the results (both using clean reads and MAGs) obtained in our study from COG and KEGG databases, the microbial functional profiles of the three samples confirm that the pathways associated with the metabolism of carbohydrates, energy, and amino acids are effectively the key pathways identified in any lignocellulosic composting, particularly in the thermophilic phase. During this phase of aerobic composting, a significant proportion of the microbial community acts on the decomposition of lignocellulose through carbohydrate and energy metabolisms. The fact that they are more active at this phase is due to the ability of microorganisms to quickly and easily degrade readily degradable carbohydrates from cellulose and hemicellulose (Kong et al. 2020; Zhang et al. 2020; Liu et al. 2022). In addition, the high abundance of metabolic functions of amino acids is triggered by the high metabolic intensity that characterizes the thermophilic phase, since amino acids provide the energy and carbon contents required for the growth and performance of microorganisms and can be used as feedstock for the synthesis of humic acids (Kong et al. 2020; Zhang et al. 2020, 2022; Li et al. 2021). The KEGG annotation also revealed a considerable number of genes associated with xenobiotic biodegradation and metabolism. The incidence of these complex compounds may result from the degradation of lignin at a later stage of the thermophilic phase (Zhang et al. 2022). In fact, the fast metabolism of amino acids, carbohydrates, and even lipids caused by high temperatures at this stage may have made available the energy and carbon content required to degrade the xenobiotic compounds (Kong et al. 2020).
The analysis of CAZymes present in our samples, considering both the clean reads and the MAGs, showed that the compost samples were enriched in GHs and GTs, as previously reported for other compost samples (Chang et al. 2022). Furthermore, the TF sample had a lower number of CDSs assigned as CAZymes, than the LP and GN samples. The same was observed when analyzing MAGs, since the LP and GN MAGs displayed a higher number of lignocellulolytic enzymes than the TF MAGs. GTs catalyze the formation of glycosidic bonds being involved in the biosynthesis of oligosaccharides, polysaccharides, and glycoconjugates (Breton et al. 2006). Among the different families, the GT2 family, which comprises enzymes derived from different sources and organisms that play a wide collection of functions (Breton et al. 2006), including cellulose/chitin synthase functions, is the most abundant. GT2 was also the main GT family represented in composting containing wheat straw, chicken manure, peanut meal, and gypsum, also in thermophilic conditions like our samples (Chang et al. 2022). However, in the context of lignocellulosic biomass conversion, GHs, which are responsible for the hydrolysis/transglycosylation of glycosidic bonds, are the most interesting enzymes as they present several catalytic activities including cellulases, endo-hemicellulases, debranching enzymes, and oligosaccharide-degrading enzymes that contribute to the composting process (Gong et al. 2020; Reichart et al. 2021). The GH13 family stood out in this study as being clearly the most abundant in the three samples. This family is the largest GH family and contains enzymes (e.g., hydrolases, transglycosidases, isomerases) exhibiting a broad range of substrate specificity and activities (Stam et al. 2006), from which the starch-degrading enzymes stand out for their potential for lignocellulose degradation (Gong et al. 2020). Other abundant GH families in the three samples were GH3, GH5, GH9, and GH31. Of note, GH3, GH5, and GH9 are among the main GH families responsible for lignocellulosic biomass degradation (Ezeilo et al. 2017), highlighting the great potential of the compost metagenomes herein presented for this process. Indeed, GH3 enzymes exhibit a vast diversity of hydrolytic activities (e.g., β-glucosidase, α-arabinofuranosidase, β-xylopyranosidase, and N-acetyl-β-glucosaminidase) that allow them to promote, among others, cellulosic biomass degradation and plant cell wall remodeling. GH5 and GH9 are important cellulase families that can act upon plant polysaccharides and carboxymethylcellulose, including endo- and exo-glucanases, endo- and exo-mannanases, β-glucosidases, and β-mannosidases (Ezeilo et al. 2017). Cellulases from the families GH5 and GH9 were the most represented in the analysis of MAGs. In a study that analyzed 60 MAGs recovered from two thermophilic composting cells, GH5 and GH9 were among the most represented cellulases in the top six degraders (Braga et al. 2021). GH5, GH8, and GH9 cellulose-degrading enzymes were also among the most abundant biocatalysts in the thermophilic compost studied by Chang et al. (2022).
The comparison study involving compost samples from the literature revealed a similar trend to the one obtained with our samples. The oligosaccharide-degrading enzymes from the GH3 family and the GH5 and GH9 cellulases were the most abundant in all cases, independently on the compost composition and process conditions, which attests again the great richness in lignocellulosic enzymes of composting environments. Importantly, the GHs’ percentage (Montella et al. 2017) and most abundant families (Allgaier et al. 2010; Kanokratana et al. 2018) previously reported were similar to those found in our study, proving the reliability of the pipeline herein implemented.
The taxonomic classification of the identified CAZymes was well aligned with the compost sample taxonomy as Proteobacteria and Bacteroidetes stood out as having the higher number of CDSs annotated as CAZymes. This evidence indicates that the taxonomic profile of the composting is directly related with its functional traits. An interesting abundance of CAZymes was also found in MAGs belonging to Actinobacteria, Balneolaeota, and Deinococcus-Thermus. Identification of CAZymes from Proteobacteria, Bacteroidetes, and Actinobacteria has been also reported in several composting samples such as those based on apple pomace (Zhou et al. 2017); on leaf and wood chip compost enriched in wheat straw, poplar, and Miscanthus (Heiss-Blanquet et al. 2016); and on shredded tree branches, leaves, grass, manure, beddings, and zoo animals’ food residues (Antunes et al. 2016). Actinomycetales appear to be common in the thermophilic and mature stages of the composting process, and CDSs annotated as lignocellulolytic enzymes belonging to this order have been reported before (Simmons et al. 2014; Antunes et al. 2016; Wang et al. 2016). Accordingly, a metatranscriptomic study on compost-derived microbial communities enriched on rice straw performed under thermophilic and mesophilic conditions revealed that lignocellulose-degrading enzymes mostly belonged to Proteobacteria and Bacteroidetes in the mesophilic microbial community, while the thermophilic was dominated by Actinobacteria (Simmons et al. 2014). Interestingly, members of Proteobacteria have also been found in samples retrieved in the end of the composting process, including Enterobacteriales and Pseudomonadales, contributing to the degradation of the remaining biomass (Antunes et al. 2016). CAZymes belonging to Balneolaeota were identified only in the TF sample. A Balneolaeota MAG predicted to have over 10 GH13 genes was recovered from a saltern pond sample with 7.5% salinity (Kimbrel et al. 2018), reinforcing again the possibility of the higher salinity of this sample in comparison with the other two under study. Lignocellulose-degrading enzymes were also found in a Deinococcus-Thermus MAG recovered from the GN sample. β-Glucosidases belonging to this phylum were also found in cow manure and rice straw composting at the thermophilic phase, which is in good agreement with the extremophile nature of the members of this phylum (Zang et al. 2017).
The experimental screening for β-glucosidase activity validated our bioinformatic pipeline as it confirmed the great abundance of positive clones exhibiting this catalytic activity, generally associated to enzymes belonging to the GH1, GH3, GH5, GH9, and GH30 families (Cairns and Esen 2010). Indeed, our samples showed great abundance of GHs within CAZymes, specifically the GH3, GH5, and GH9 families, as aforementioned. Since β-glucosidases are generally the rate-limiting enzymes in the cellulose degradation (Zang et al. 2018), the identified enzymes, which are likely to be active at high temperatures considering the thermophilic origin of the compost, can represent a promising enzymatic pool to be explored for cellulose degradation in plant biomass under harsh industrial conditions. The lower percentage obtained in the experimental approach in comparison with the bioinformatic prediction may be explained by the need for optimizing the growth conditions, the catalytic activity, and, probably, the host itself. In fact, the screening was performed in standard conditions of temperature and incubation times, and further optimization may render a similar percentage to the ones predicted in silico.
In conclusion, the taxonomy and functional profile of the compost samples’ microbiome was successfully characterized using the developed bioinformatic pipeline, which was validated using previously reported data. Bacteria clearly dominated all samples, with the classes Gammaproteobacteria, Alphaproteobacteria, and Balneolia being the most abundant. These data suggest that bacterial enzymatic activity is the main driver of lignocellulose degradation in the thermophilic compost samples herein studied. The analysis of our samples together with samples retrieved from the literature confirmed that composting, independently of the composition and process conditions, is a promising source of lignocellulolytic enzymes, especially GHs, which goes well in line with one of the most abundant functional categories predicted in our samples (carbohydrate transport and metabolism). Indeed, around 1% of the CDSs of the compost metagenomes were predicted to be from putative CAZymes. Specifically within GHs, our samples were enriched in cellulases from the GH5 and GH9 families and oligosaccharide-degrading enzymes from the GH3 family. These families are known to contain β-glucosidases, whose catalytic activity was further validated by our experimental functional screening. As our samples were retrieved from the thermophilic phase of the composting process, it is expected that at least a considerable proportion of the identified bacterial GHs would be resistant to harsh conditions, namely, high temperatures. Therefore, they could be considered promising biocatalysts for developing efficient and sustainable strategies for lignocellulosic biomass conversion.
Data availability
The authors confirm that the datasets supporting the findings and conclusions of this study are available within the article and its supplemental information file. Additional data can be provided upon request.
References
Alfredson GA, Kristjansson JK, Sigridur H, Stetter KO (1988) Rhodothermus marinus, gen. nov., sp. nov., a thermophibic, halophilic bacterium from submarine hot springs in Iceland. J Gen Microbiol 134:299–306
Ali N, Zhang Q, Liu Z-Y, Li F-L, Lu M, Fang X-C (2020) Emerging technologies for the pretreatment of lignocellulosic materials for bio-based products. Appl Microbiol Biotechnol 104:455–473. https://doi.org/10.1007/s00253-019-10158-w
Allgaier M, Reddy A, Park JI, Ivanova N, D’Haeseleer P, Lowry S, Sapra R, Hazen TC, Simmons BA, Vandergheynst JS, Hugenholtz P (2010) Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One 5:e8812. https://doi.org/10.1371/journal.pone.0008812
Almeida E, Carvalho MF, Lage OM (2022) Culturomics remains a highly valuable methodology to obtain rare microbial diversity with putative biotechnological potential from two Portuguese salterns. Front Biosci - Elit 14:11. https://doi.org/10.31083/j.fbe1402011
Alneberg J, Bjarnason BS, De Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C (2014) Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. https://doi.org/10.1038/nmeth.3103
Antunes LP, Martins LF, Pereira RV, Thomas AM, Barbosa D, Lemos LN, Silva GMM, Moura LMS, Epamino GWC, Digiampietri LA, Lombardi KC, Ramos PL, Quaggio RB, De Oliveira JCF, Pascon RC, Da Cruz JB, Da Silva AM, Setubal JC (2016) Microbial community structure and dynamics in thermophilic composting viewed through metagenomics and metatranscriptomics. Sci Rep 6:38915. https://doi.org/10.1038/srep38915
Awasthi MK, Pandey AK, Khan J, Bundela PS, Wong JWC, Selvam A (2014) Evaluation of thermophilic fungal consortium for organic municipal solid waste composting. Bioresour Technol 168:214–221. https://doi.org/10.1016/j.biortech.2014.01.048
Ballardo C, del Vargas-García MC, Sánchez A, Barrena R, Artola A (2020) Adding value to home compost: biopesticide properties through Bacillus thuringiensis inoculation. Waste Manag 106:32–43. https://doi.org/10.1016/j.wasman.2020.03.003
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Braga LPP, Pereira RV, Martins LF, Moura LMS, Sanchez FB, Patané JSL, da Silva AM, Setubal JC (2021) Genome-resolved metagenome and metatranscriptome analyses of thermophilic composting reveal key bacterial players and their metabolic interactions. BMC Genomics 22:1–19. https://doi.org/10.1186/s12864-021-07957-9
Breton C, Šnajdrová L, Jeanneau C, Koča J, Imberty A (2006) Structures and mechanisms of glycosyltransferases. Glycobiology 16:29R-37R. https://doi.org/10.1093/glycob/cwj016
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. https://doi.org/10.1038/nmeth.3176
Cairns JRK, Esen A (2010) β-Glucosidases. Cell Mol Life Sci 67:3389–3405. https://doi.org/10.1007/s00018-010-0399-2
Cantarel BI, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. https://doi.org/10.1093/nar/gkn663
Cao L, Liao L, Su C, Mo T, Zhu F, Qin R, Li R (2021) Metagenomic analysis revealed the microbiota and metabolic function during co-composting of food waste and residual sludge for nitrogen and phosphorus transformation. Sci Total Environ 773:145561. https://doi.org/10.1016/j.scitotenv.2021.145561
Chang W, Feng W, Yang Y, Shen Y, Song T, Li Y, Cai W (2022) Metagenomics analysis of the effects of Agaricus bisporus mycelia on microbial diversity and CAZymes in compost. PeerJ 10:1–21. https://doi.org/10.7717/peerj.14426
Chin KL, H’ng PS, Chai EW, Khoo PS, Lee CL, Go WZ (2020) Valorization of lignocellulosic food industry waste in Malaysia by accelerated co-composting method: changes in physicochemical and microbial community. Waste Biomass Valor 11:4871–4884. https://doi.org/10.1007/s12649-019-00825-4
Costa ÂMA, Santos AO, Sousa J, Rodrigues JL, Gudiña EJ, Silvério SC, Rodrigues LR (2021) Improved method for the extraction of high-quality DNA from lignocellulosic compost samples for metagenomic studies. Appl Microbiol Biotechnol 105:8881–8893. https://doi.org/10.1007/s00253-021-11647-7
Datta S, Rajnish KN, Samuel MS, Pugazlendhi A, Selvarajan E (2020) Metagenomic applications in microbial diversity, bioremediation, pollution monitoring, enzyme and drug discovery. A Review. Environ Chem Lett 18:1229–1241. https://doi.org/10.1007/s10311-020-01010-z
DeCastro ME, Rodríguez-Belmonte E, González-Siso MI (2016) Metagenomics of thermophiles with a focus on discovery of novel thermozymes. Front Microbiol 7:1521. https://doi.org/10.3389/fmicb.2016.01521
Devi A, Bajar S, Kour H, Kothari R, Pant D, Singh A (2022) Lignocellulosic biomass valorization for bioethanol production: a circular bioeconomy approach. Bioenergy Res 15:1820–1841. https://doi.org/10.1007/s12155-022-10401-9
Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, Terrapon N (2022) The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res 50:D571–D577. https://doi.org/10.1093/nar/gkab1045
Duan Y, Tarafdar A, Kumar V, Ganeshan P, Rajendran K, Shekhar Giri B, Gómez-García R, Li H, Zhang Z, Sindhu R, Binod P, Pandey A, Taherzadeh MJ, Sarsaiya S, Jain A, Kumar Awasthi M (2022) Sustainable biorefinery approaches towards circular economy for conversion of biowaste to value added materials and future perspectives. Fuel 325:124846. https://doi.org/10.1016/j.fuel.2022.124846
Ezeilo UR, Zakaria II, Huyop F, Wahab RA (2017) Enzymatic breakdown of lignocellulosic biomass: the role of glycosyl hydrolases and lytic polysaccharide monooxygenases. Biotechnol Biotechnol Equip 31:647–662. https://doi.org/10.1080/13102818.2017.1330124
Finore I, Feola A, Russo L, Cattaneo A, Di Donato P, Nicolaus B, Poli A, Romano I (2023) Thermophilic bacteria and their thermozymes in composting processes: a review. Chem Biol Technol Agric 10:7. https://doi.org/10.1186/s40538-023-00381-z
Gomes I, Gomes J, Steiner W (2003) Highly thermostable amylase and pullulanase of the extreme thermophilic eubacterium Rhodothermus marinus: production and partial characterization. Bioresour Technol 90:207–214. https://doi.org/10.1016/S0960-8524(03)00110-X
Gong G, Zhou S, Luo R, Gesang Z, Suolang S (2020) Metagenomic insights into the diversity of carbohydrate-degrading enzymes in the yak fecal microbial community. BMC Microbiol 20:302. https://doi.org/10.1186/s12866-020-01993-3
Goyal S, Dhull SK, Kapoor KK (2005) Chemical and biological changes during composting of different organic wastes and assessment of compost maturity. Bioresour Technol 96:1584–1591. https://doi.org/10.1016/j.biortech.2004.12.012
Guo R, Li G, Jiang T, Schuchardt F, Chen T, Zhao Y, Shen Y (2012) Effect of aeration rate, C/N ratio and moisture content on the stability and maturity of compost. Bioresour Technol 112:171–178. https://doi.org/10.1016/j.biortech.2012.02.099
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Hahnke RL, Meier-Kolthoff JP, García-López M, Mukherjee S, Huntemann M, Ivanova NN, Woyke T, Kyrpides NC, Klenk HP, Göker M (2016) Genome-based taxonomic classification of Bacteroidetes. Front Microbiol 7:2003. https://doi.org/10.3389/fmicb.2016.02003
Hassan SS, Williams GA, Jaiswal AK (2019) Moving towards the second generation of lignocellulosic biorefineries in the EU: Drivers, challenges, and opportunities. Renew Sustain Energy Rev 101:590–599. https://doi.org/10.1016/j.rser.2018.11.041
Hayat R, Sheirdil RA, Iftikhar-Ul-Hassan M, Ahmed I (2013) Characterization and identification of compost bacteria based on 16S rRNA gene sequencing. Ann Microbiol 63:905–912. https://doi.org/10.1007/s13213-012-0542-4
He X, Li N, Chen X, Zhang Y, Zhang X, Song X (2020) Pedobacter indicus sp. nov., isolated from deep-sea sediment. Antonie Van Leeuwenhoek 113:357–364. https://doi.org/10.1007/s10482-019-01346-9
Heiss-Blanquet S, Fayolle-Guichard F, Lombard V, Hébert A, Coutinho PM, Groppi A, Barre A, Henrissat B (2016) Composting-like conditions are more efficient for enrichment and diversity of organisms containing cellulase-encoding genes than submerged cultures. PLoS ONE 11:1–22. https://doi.org/10.1371/journal.pone.0167216
Hemati A, Aliasgharzad N, Khakvar R, Delangiz N, Asgari Lajayer B, van Hullebusch ED (2022) Bioaugmentation of thermophilic lignocellulose degrading bacteria accelerate the composting process of lignocellulosic materials. Biomass Convers Biorefinery 1–15. https://doi.org/10.1007/s13399-021-02238-7
Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P (2008) eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res 36:D250–D254. https://doi.org/10.1093/nar/gkm796
Jo G, Hong S, Kim H, Zhuliping, Ahn D (2021) Removal of biological organics in high-salinity wastewater produced from methylcellulose production and subsequent changes in the microbial community. Environ Eng Res 26:200187. https://doi.org/10.4491/eer.2020.187
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z (2019) MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. https://doi.org/10.7717/peerj.7359
Kanokratana P, Wongwilaiwalin S, Mhuantong W, Tangphatsornruang S, Eurwilaichitr L, Champreda V (2018) Characterization of cellulolytic microbial consortium enriched on Napier grass using metagenomic approaches. J Biosci Bioeng 125:439–447. https://doi.org/10.1016/j.jbiosc.2017.10.014
Kaushal G, Kumar J, Sangwan RS, Singh SP (2018) Metagenomic analysis of geothermal water reservoir sites exploring carbohydrate-related thermozymes. Int J Biol Macromol 119:882–895. https://doi.org/10.1016/j.ijbiomac.2018.07.196
Kimbrel JA, Ballor N, Wu YW, David MM, Hazen TC, Simmons BA, Singer SW, Jansson JK (2018) Microbial community structure and functional potential along a hypersaline gradient. Front Microbiol 9:1–15. https://doi.org/10.3389/fmicb.2018.01492
Kong W, Sun B, Zhang J, Zhang Y, Gu L, Bao L, Liu S (2020) Metagenomic analysis revealed the succession of microbiota and metabolic function in corncob composting for preparation of cultivation medium for Pleurotus ostreatus. Bioresour Technol 306:123156. https://doi.org/10.1016/j.biortech.2020.123156
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Kulikowska D, Bernat K, Zaborowska M, Zielińska M (2022) Municipal sewage sludge composting in the two-stage system: the role of different bulking agents and amendments. Energies 15:5014. https://doi.org/10.3390/en15145014
Le TTH, Nguyen TB, Nguyen HD, Nguyen HD, Le NG, Dao TK, Nguyen TQ, Do TH, Truong NH (2022) De novo metagenomic analysis of microbial community contributing in lignocellulose degradation in humus samples harvested from Cuc Phuong tropical forest in Vietnam. Diversity 14:220. https://doi.org/10.3390/d14030220
Li C, Lai Q, Fu Y, Chen S, Shao Z (2013) Galbibacter marinus sp. nov., isolated from deep-sea sediment. Int J Syst Evol Microbiol 63:1427–1430. https://doi.org/10.1099/ijs.0.044305-0
Li D, Liu CM, Luo R, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. https://doi.org/10.1093/bioinformatics/btv033
Li G, Zhu Q, Niu Q, Meng Q, Yan H, Wang S, Li Q (2021) The degradation of organic matter coupled with the functional characteristics of microbial community during composting with different surfactants. Bioresour Technol 321:124446. https://doi.org/10.1016/j.biortech.2020.124446
Liu Q, He X, Luo G, Wang K, Li D (2022) Deciphering the dominant components and functions of bacterial communities for lignocellulose degradation at the composting thermophilic phase. Bioresour Technol 348:126808. https://doi.org/10.1016/j.biortech.2022.126808
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. https://doi.org/10.1093/nar/gkt1178
Ma C, Lo PK, Xu J, Li M, Jiang Z, Li G, Zhu Q, Li X, Leong SY, Li Q (2020) Molecular mechanisms underlying lignocellulose degradation and antibiotic resistance genes removal revealed via metagenomics analysis during different agricultural wastes composting. Bioresour Technol 314:123731. https://doi.org/10.1016/j.biortech.2020.123731
Madhavan A, Sindhu R, Parameswaran B, Sukumaran RK, Pandey A (2017) Metagenome analysis : a powerful tool for enzyme bioprospecting. Appl Biochem Biotechnol 183:636–651. https://doi.org/10.1007/s12010-017-2568-3
Martins I, Garcia H, Varela A, Núñez O, Planchon S, Galceran MT, Renaut J, Rebelo LPN, Silva Pereira C (2014) Investigating Aspergillus nidulans secretome during colonisation of cork cell walls. J Proteomics 98:175–188. https://doi.org/10.1016/j.jprot.2013.11.023
Menzel P, Ng KL, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. https://doi.org/10.1038/ncomms11257
Montella S, Ventorino V, Lombard V, Henrissat B, Pepe O, Faraco V (2017) Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci Rep 7:42623. https://doi.org/10.1038/srep42623
Palaniveloo K, Amran MA, Norhashim NA, Mohamad-Fauzi N, Peng-Hui F, Hui-Wen L, Kai-Lin Y, Jiale L, Chian-Yee MG, Jing-Yi L, Gunasekaran B, Razak SA (2020) Food waste composting and microbial community structure profiling. Processes 8:723. https://doi.org/10.3390/pr8060723
Reichart NJ, Bowers RM, Woyke T, Hatzenpichler R (2021) High potential for biomass-degrading enzymes revealed by hot spring metagenomics. Front Microbiol 12:668238. https://doi.org/10.3389/fmicb.2021.668238
Sánchez ÓJ, Ospina DA, Montoya S (2017) Compost supplementation with nutrients and microorganisms in composting process. Waste Manag 69:136–153. https://doi.org/10.1016/j.wasman.2017.08.012
Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF (2018) Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3:836–843. https://doi.org/10.1038/s41564-018-0171-1
Simmons CW, Reddy AP, D’haeseleer P, Khudyakov J, Billis K, Pati A, Simmons BA, Singer SW, Thelen MP, Vandergheynst JS (2014) Metatranscriptomic analysis of lignocellulolytic microbial communities involved in high-solids decomposition of rice straw. Biotechnol Biofuels 7:1–13. https://doi.org/10.1186/s13068-014-0180-0
Song C, Li M, Xi B, Wei Z, Zhao Y, Jia X, Qi H, Zhu C (2015) Characterisation of dissolved organic matter extracted from the bio-oxidative phase of co-composting of biogas residues and livestock manure using spectroscopic techniques. Int Biodeterior Biodegrad 103:38–50. https://doi.org/10.1016/j.ibiod.2015.03.032
Sousa J, Silvério SC, Costa AMA, Rodrigues LR (2022) Metagenomic approaches as a tool to unravel promising biocatalysts from natural resources: soil and water. Catalysts 12:385. https://doi.org/10.3390/catal12040385
Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B (2006) Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel 19:555–562. https://doi.org/10.1093/protein/gzl044
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. https://doi.org/10.1093/nar/28.1.33
Tian W, Sun Q, Xu D, Zhang Z, Chen D, Li C, Shen Q, Shen B (2013) Succession of bacterial communities during composting process as detected by 16S rRNA clone libraries analysis. Int Biodeterior Biodegrad 78:58–66. https://doi.org/10.1016/j.ibiod.2012.12.008
Unsal T, Sozudogru Ok S (2001) Description of characteristics of humic substances from different waste materials. Bioresour Technol 78:239–242. https://doi.org/10.1016/S0960-8524(01)00019-0
Urakami T, Tamaoka J, Suzuki KI, Komagata K (1989) Paracoccus alcaliphilus sp. nov., an alkaliphilic and facultatively methylotrophic bacterium. Int J Syst Bacteriol 39:116–121. https://doi.org/10.1099/00207713-39-2-116
Usmani Z, Sharma M, Awasthi AK, Lukk T, Tuohy MG, Gong L, Nguyen-Tri P, Goddard AD, Bill RM, Nayak SC, Gupta VK (2021) Lignocellulosic biorefineries: the current state of challenges and strategies for efficient commercialization. Renew Sustain Energy Rev 148:111258. https://doi.org/10.1016/j.rser.2021.111258
Wang C, Dong D, Wang H, Müller K, Qin Y, Wang H, Wu W (2016) Metagenomic analysis of microbial consortia enriched from compost: new insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol Biofuels 9:22. https://doi.org/10.1186/s13068-016-0440-2
Wang W, Zhang Q, Sun X, Chen D, Insam H, Koide RT, Zhang S (2020) Effects of mixed-species litter on bacterial and fungal lignocellulose degradation functions during litter decomposition. Soil Biol Biochem 141:107690. https://doi.org/10.1016/j.soilbio.2019.107690
Wang ZJ, Liu QQ, Zhao LH, Du ZJ, Chen GJ (2015) Bradymonas sediminis gen. nov., sp. nov., isolated from coastal sediment, and description of Bradymonadaceae fam. nov. and Bradymonadales ord. nov. Int J Syst Evol Microbiol 65:1542–1549. https://doi.org/10.1099/ijs.0.000135
Wu YW, Simmons BA, Singer SW (2016) MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. https://doi.org/10.1093/bioinformatics/btv638
Yassin AF, Chen WM, Hupfer H, Siering C, Kroppenstedt RM, Arun AB, Lai WA, Shen FT, Rekha PD, Young CC (2007) Lysobacter defluvii sp. nov., isolated from municipal solid waste. Int J Syst Evol Microbiol 57:1131–1136. https://doi.org/10.1099/ijs.0.64966-0
Zang X, Liu M, Fan Y, Xu J, Xu X, Li H (2018) The structural and functional contributions of β-glucosidase-producing microbial communities to cellulose degradation in composting. Biotechnol Biofuels 11:51. https://doi.org/10.1186/s13068-018-1045-8
Zang X, Liu M, Wang H, Fan Y, Zhang H, Liu J, Xing E, Xu X, Li H (2017) The distribution of active β-glucosidase-producing microbial communities in composting. Can J Microbiol 63:998–1008. https://doi.org/10.1139/cjm-2017-0368
Zhang WM, Yu CX, Wang XJ, Hai L (2020) Increased abundance of nitrogen transforming bacteria by higher C/N ratio reduces the total losses of N and C in chicken manure and corn stover mix composting. Bioresour Technol 297:122410. https://doi.org/10.1016/j.biortech.2019.122410
Zhang Y, Chen M, Guo J, Liu N, Yi W, Yuan Z, Zeng L (2022) Study on dynamic changes of microbial community and lignocellulose transformation mechanism during green waste composting. Eng Life Sci 22:376–390. https://doi.org/10.1002/elsc.202100102
Zhou M, Guo P, Wang T, Gao L, Yin H, Cai C, Gu J, Lü X (2017) Metagenomic mining pectinolytic microbes and enzymes from an apple pomace-adapted compost microbial community. Biotechnol Biofuels 10:198. https://doi.org/10.1186/s13068-017-0885-y
Zhu W, Lomsadze A, Borodovsky M (2010) Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. https://doi.org/10.1093/nar/gkq275
Acknowledgements
The authors acknowledge the Portuguese composting units Terra Fértil, Lipor, and Gintegral for kindly supplying the compost samples. CSP acknowledges her research grant UMINHO/BID/2021/12, under the scope of the B3iS project (PTDC/BII-BIO/5554/2020). JS acknowledges the PhD fellowship (2022.11695.BD) funded by the Portuguese Foundation for Science and Technology (FCT).
Funding
Open access funding provided by FCT|FCCN (b-on). This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UIDB/04469/2020 unit, the projects LIGNOZYMES—Metagenomics approach to unravel the potential of lignocellulosic residues towards the discovery of novel enzymes (POCI-01–0145-FEDER-029773), and B3iS—Biodiversity and Bioprospecting of Biosurfactants in Saline Environments (PTDC/BII-BIO/5554/2020); and by RNCA Advanced Computing Project MetaLignoZymes, metagenomic analysis of lignocellulosic residues towards the discovery of novel enzymes (CPCA/A0/408464/2021).
Author information
Authors and Affiliations
Contributions
JS, AC, and AS conducted the experimental work. CSP, RFD, TR, and PS developed the bioinformatic pipeline and performed the respective analysis. CSP, JS, AC, AS, SS, and LR conceived and designed the research. SS, RFD, and LR obtained financial support and coordinated the research. CSP, JS, RFD, and SS wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This work does not contain any studies with human participants or animals.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Santos-Pereira, C., Sousa, J., Costa, Â.M.A. et al. Functional and sequence-based metagenomics to uncover carbohydrate-degrading enzymes from composting samples. Appl Microbiol Biotechnol 107, 5379–5401 (2023). https://doi.org/10.1007/s00253-023-12627-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12627-9