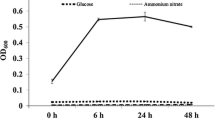
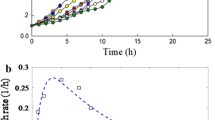
Abstract
The Arthrobacter sp. strain AK-YN10 is an s-triazine pesticide degrading bacterium isolated from a sugarcane field in Central India with history of repeated atrazine use. AK-YN10 was shown to degrade 99 % of atrazine in 30 h from media supplemented with 1000 mg L−1 of the herbicide. Draft genome sequencing revealed similarity to pAO1, TC1, and TC2 catabolic plasmids of the Arthrobacter taxon. Plasmid profiling analyses revealed the presence of four catabolic plasmids. The trzN, atzB, and atzC atrazine-degrading genes were located on a plasmid of approximately 113 kb.
The flagellar operon found in the AK-YN10 draft genome suggests motility, an interesting trait for a bioremediation agent, and was homologous to that of Arthrobacter chlorophenolicus. The multiple s-triazines degradation property of this isolate makes it a good candidate for bioremediation of soils contaminated by s-triazine pesticides.



Similar content being viewed by others
References
Anderson DG, Mckay LL (1983) Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Microbiol Biotechnol 46:549–552
Arbeli Z, Fuentes C (2010) Prevalence of the gene trzN and biogeographic patterns among atrazine-degrading bacteria isolated from 13 Colombian agricultural soils. FEMS Microbiol Ecol 73:611–623
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75
Cai B, Han Y, Liu B, Ren Y, Jiang S (2003) Isolation and characterization of an atrazine-degrading bacterium from industrial waste water in China. Lett Appl Microbiol 36:272–276
De Souza ML, Wackett LP, Sadowski MJ (1998) The atzABC genes encoding atrazine catabolism are located on a self transmissible plasmid in Pseudomonas sp. strain ADP. Appl Environ Microbiol 64:2323–2326
Devers M, Soulas G, Martin-Laurent F (2004) Real-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J Microbiol Methods 56:1–3
Devers M, Azhari NE, Kolic NU, Martin-Laurent F (2007) Detection and organization of atrazine-degrading genetic potential of seventeen bacterial isolates belonging to divergent taxa indicate a recent common origin of their catabolic functions. FEMS Microbiol Lett 273:78–86
Eckhardt T (1978) A rapid method for identification of plasmid deoxyribonucleic acid in bacteria. Plasmid 1:584–588
Farre MJ, Franch MI, Malato S, Ayllon JA, Peral J, Domenech X (2005) Degradation of some biorecalcitrant pesticides by homogeneous and heterogeneous photocatalytic ozonation. Chemosphere 58:1127–1133
Fenner K, Canonica S, Wackett LP, Elsner M (2013) Evaluating pesticide degradation in the environment: blind spots and emerging opportunities. Science 341:752–758
Fruchey I, Shapir N, Sadowsky MJ, Wackett LP (2003) On the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP. Appl Environ Microbiol 69:3653–3657
Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:181–184
Hayes TB, Haston K, Tsui M, Hoang A, Haeffele C, Vonk A (2002) Feminization of male frogs in the wild. Nature 419:895–896
Hayes TB, Khoury V, Narayan A, Nazir M, Park A, Brown T, Adame L, Chan E, Buchholz D, Stueve T, Gallipeau S (2010) Atrazine induces complete feminization and chemical castration in male African clawed frogs (Xenopus laevis). Proc Natl Acad Sci U S A 107:4612–4617
Igloi GL, Brandsch R (2003) Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J Bacteriol 185:1976–1986
Jerke K, Nakatsu CH, Beasley F, Konopka A (2008) Comparative analysis of eight Arthrobacter plasmids. Plasmid 59:73–85
Kallimanis A, Labutti KM, Lapidus A, Clum A, Lykidis A, Mavromatis K, Pagani I, Liolios K, Ivanova N, Goodwin L, Pitluck S, Chen A, Palaniappan K, Markowitz V, Bristow J, Velentzas AD, Perisynakis A, Ouzounis CC, Kyrpides NC, Koukkou AI, Drainas C (2011) Complete genome sequence of Arthrobacter phenanthrenivorans type strain (Sphe3). Stand Genomic Sci 29:123–130
Kapley A, Tolamare A, Purohit HJ (2001) Role of oxygen in partial utilization of phenol in continuous culture. World J Microbiol Biotechnol 17:801–804
Krutz LJ, Shaner DL, Weaver MA, Webb RM, Zablotowicz RM, Reddy KN, Huang Y, Thomson SJ (2010) Agronomic and environmental implications of enhanced s-triazine degradation. Pest Manag Sci 66:461–481
Lovley DR (2003) Cleaning up with genomics: Applying molecular biology to Bioremediation. Nat Rev Microbiol 1:35–44
Michael CR, Alavanja PH (2009) Pesticides use and exposure extensive worldwide. Rev Environ Health 24:303–309
Mongodin EF, Shapir N, Daugherty SC, Deboy RT, Emerson JB, Shvartzbeyn A, Radune D, Vamathevan J, Riggs F, Grinberg V, Khouri H, Wackett LP, Nelson KE, Sadowsky MJ (2006) Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. Plos Genet 2:2094–2106
Mulbry WW, Zhu H, Nour SM, Topp E (2002) The triazine hydrolase gene trzN from Nocardioides sp. strain C190: Cloning and construction of gene-specific primers. FEMS Microbiol Lett 206:75–79
Niewerth H, Schuldes J, Parschat K, Kiefer P, Vorholt JA, Daniel R, Fetzner S (2013) Complete genome sequence and metabolic potential of the quinaldine-degrading bacterium Arthrobacter sp. Rue61a. BMC Genomics 13:534
Nordin K, Unell M, Jansson JK (2005) Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl Environ Microbiol 71:6538–6544
Piutti S, Semon E, Landry D, Hartmann A, Dousset S, LichtfouseE TE, Soulas G, Martin-Laurent F (2003) Isolation and characterization of Nocardioides sp. SP12, an atrazine degrading bacterial strain possessing the gene trzN frombulk- and maize rhizosphere soil. FEMS Microbiol Lett 221:111–117
Sagarkar S, Mukherjee S, Nousiainen A, Björklöf K, Purohit HJ, Jørgensen KS, Kapley A (2013) Monitoring bioremediation of atrazine in soil microcosms using molecular tools. Environ Pollut 172:108–115
Sagarkar S, Bhardwaj P, Yadav T, Qureshi A, Khardenavis A, Purohit HJ, Kapley A (2014a) Draft genome of atrazine-utilizing bacteria isolated from Indian agricultural soil. Genome Announc 2:e01149–13
Sagarkar S, Nousiainen A, Shaligram S, Björklöf K, Lindström K, Jørgensen KS, Kapley A (2014b) Soil mesocosm studies on atrazine bioremediation. J Environ Manage 139:208–216
Sajjaphan K, Shapir N, Wackett LP, Palmer M, Blackmon B, Tomkins J, Sadowsky MJ (2004) Arthrobacter aurescens TC1 atrazine catabolism genes trzN, atzB, and atzC are linked on a 160-kilobase region and are functional in Escherichia coli. Appl Environ Microbiol 70:4402–4407
Sene L, Converti A, Secchi GAR, Simão RDCG (2010) New aspects on atrazine biodegradation. Braz Arch Biol Technol 53:487–496
Strong LC, Rosendahl C, Johnson G, Sadowsky MJ, Wackett LP (2002) Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds. Appl Environ Microbiol 68:5973–5980
Topp E, Mulbry WM, Zhu H, Nour SM, Cuppels D (2000) Characterization of s-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. Appl Environ Microbiol 66:3134–3141
Udiković-Kolić N, Martin-Laurent F, Devers M, Petrić I, Kolar AB, Hršak D (2008) Genetic potential, diversity and activity of an atrazine degrading community enriched from a herbicide factory effluent. J Appl Microbiol 105:1334–1343
Udiković-Kolić N, Scott C, Martin-Laurent F (2012) Evolution of atrazine-degrading capabilities in the environment. Appl Microbiol Biotechnol 96:1175–1189
Vaishampayan PA, Kanekar PP, Dhakephalkar PK (2007) Isolation and characterization of Arthrobacter sp. strain MCM B-436, an atrazine-degrading bacterium, from rhizospheric soil. Int Biodeterior Biodegrad 60:273–278
Wheatcroft R, McRae DG, Miller RW (1990) Changes in the Rhizobium meliloti genome and the ability to detect supercoiled plasmids during bacteroid development. Mol Plant-Microbe Interact 3:9–17
Yamazaki K, Fujii K, Iwasaki A, Takagi K, Satsuma K, Harada N, Uchimura T (2008) Different substrate specificities of two triazine hydrolases (TrzNs) from Nocardioides species. FEMS Microbiol Lett 286:171–177
Acknowledgments
Funds from the Department of Biotechnology, New Delhi are gratefully acknowledged. Author Sneha Sagarkar acknowledges the Raman Charpak funding to work in the laboratory of Agroecology at INRA of Dijon (France). Pooja Bhardwaj is grateful to CSIR/UGC (India) for a senior research fellowship award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Compliance with Ethical Standards
ᅟ
Conflict of Interest
All authors declare that they have no competing interests.
Ethical approval
No ethical approval is required since this study did not involve animals/human samples.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1072 kb)
Rights and permissions
About this article
Cite this article
Sagarkar, S., Bhardwaj, P., Storck, V. et al. s-triazine degrading bacterial isolate Arthrobacter sp. AK-YN10, a candidate for bioaugmentation of atrazine contaminated soil. Appl Microbiol Biotechnol 100, 903–913 (2016). https://doi.org/10.1007/s00253-015-6975-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6975-5