Abstract
Riemerella anatipestifer (RA) infections cause major economic losses in the duck industry. In this study, an immunogenic protein, chaperonin GroEL (GroEL), was identified from the outer membrane of RA strain WJ4 by immunoproteomic assay based on matrix-assisted laser desorption/ionization time of flight mass spectrometry. The complete sequence of the encoding gene, chaperonin groEL (groEL) was amplified and determined to be 1,629 base pairs in length. groEL was then cloned into expression vector pGEX-6P-1, and the expression of the recombinant GroEL (rGroEL) in Escherichia coli strain BL21 was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western blotting analysis. Immunization assay showed that ducklings or rabbits immunized with purified rGroEL generated 53- or 160-fold more anti-GroEL antibodies than those with no immunization. Importantly, bactericidal assay showed that rabbit anti-GroEL serum killed 30.0–57.3% of bacteria representing different serotypes, while rabbit anti-bacterin serum killing activity exhibits large serotype-dependent variations between 0.2% and 63.6%. Animal challenge experiment showed that ducklings immunized with rGroEL were 50%, 37.5%, and 37.5% protected from the challenge with RA strains WJ4 (serotype 1), Th4 (serotype 2), and YXb-2 (serotype 10), respectively. In addition, groEL from 34 additional RA strains was amplified by polymerase chain reaction (PCR), and products from nine were sequenced. groEL is highly conserved among RA strains, as the DNA sequence identity was over 97.5% between WJ4 and the nine additional strains. Our results suggest that GroEL may be a good candidate for new RA vaccine development.
Similar content being viewed by others
Introduction
Riemerella anatipestifer (RA), a Gram-negative, nonmotile, and nonspore forming rod-shaped bacterium, is the causative agent of an epizootic disease in poultry, especially in ducks (Pathanasophon et al. 2002). Endemic RA infections are usually restricted to commercial duck and turkey flocks, but other poultry species such as chicken and geese are also susceptible to the infection (Cooper 1989; Helfer and Helmboldt 1977). For ducks under about 8 weeks of age, RA infection is often acute, with the mortality rate usually between 10% and 30%, though mortality of as high as 75% has been recorded in infected duck farms (Subramaniam et al. 2000). Up to date, 21 serotypes of RA have been identified (Kardos et al. 2007; Pathanasophon et al. 2002). The occurrence of different serotypes has been reported in RA field cases, but serotypes 1, 2, and 10 are responsible for most of the major outbreaks in China (Hu et al. 2010).
Vaccines based on inactivated bacteria confer some protection against infection with homologous strains or serotypes, but showed no significant cross-protection against heterologous strain challenges (Sandhu 1979). Outer membrane protein A (OmpA) and a 41-kDa partial protein (P45N′) of RA have been characterized to be immunogenic proteins; however, the established subunit vaccine does not provide an effective protection against heterologous strain challenges either (Huang et al. 2002; Subramaniam et al. 2000). Immunoproteomics is a mass-spectrometry-based method to study the proteins involved in immune response (Aebersold and Mann 2003). With the significant recent advances in proteomics technology (Aebersold and Mann 2003; Han et al. 2008), immunoproteomics has become widely used as a powerful means to discover new immunogenic proteins (Zhang and Lu 2007). In this study, we identified chaperonin GroEL (GroEL) as a novel immunogenic protein of RA by immunoproteomics. GroEL of other bacteria has been reported to be a protective immunogenic protein (Sinha and Bhatnagar 2010; Khan et al. 2009). Strain WJ4 was chosen for the study because of its favorable antigenicity based on our previous study. The immunogenicity of the recombinant GroEL (rGroEL), bactericidal activity of the anti-serum, and the protection rate of the immunized ducklings were also investigated.
Materials and methods
Bacterial strains and growth conditions
Thirty-one RA strains (DY-1, CH3, CH1, WJ4, Th4, YXb12, NJ-2, CQ2, CQ3, CQ4, NJ-4, JY-4, YXb14, FXb6, Yb2, Yb3, JY-1, JY-2, NJ-3, SC1, SC2, SC3, SC4, HXb2, YXb11, YXb1, R20, YXb13, YXb15, YXD1, and JY-6) were isolated from infected duck farms in China between 1997 and 2009 (Hu et al. 2010). Four RA serotype reference strains (P2123, D26220, 8785, and D743) were donated generously by Dr. Guoqiang Zhu and Dr. Zhizhong Cui. Strain WJ4 was deposited in China General Microbiological Culture Collection Center (CGMCC no. 5264) and chosen for immunoproteomics study because of its favorable antigenicity.
RA strains were cultured on tryptic soy agar (TSA, Difco, Detroit, MI, USA) or in tryptic soy broth (TSB, Difco) at 37°C with 5% CO2. Escherichia coli strains DH5α (Invitrogen, Carlsbad, CA, USA) and BL21 (DE3, Stratagene, La Jolla, CA, USA) were grown on Luria–Bertani (LB) agar plates or broth at 37°C, and transformants were selected on media supplemented with 100 μg/mL ampicillin.
General DNA reagents and methods
Taq DNA polymerase, DNA restriction endonucleases, and T4 DNA ligase were purchased from Fermentas (Hanover, MD, USA). TA Cloning Kit was purchased from Invitrogen (Carlsbad, CA, USA). Bacterial chromosomal DNA was isolated using Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. DNA nucleotide sequences were obtained via 3070xl DNA analyzer (Applied Biosystems, Foster City, CA, USA) and analyzed with DNASTAR software (DNASTAR Inc., Madison, WI, USA). DNA manipulations and general molecular biology techniques were performed according to standard procedures (Cullen et al. 2002; Sinha et al. 2005). Oligonucleotides used in this study are listed in Table 1.
Preparation of bacterial outer membrane proteins and rabbit anti-serum against RA
The outer membrane proteins of RA strain WJ4 were extracted as described (Cullen et al. 2002; Sinha et al. 2005) with minor modifications. Briefly, bacteria were grown in TSB to mid-exponential phase (OD600 = 0.8) and centrifuged at 10,000×g for 20 min at 4°C. The pelleted bacteria were suspended in 12 mL of buffer A (80 mM Tris–HCl, pH 7.4, 1.2 M NaCl) and briefly sonicated on ice (5 s for 80 times at 400 W, with 10-s intervals between repeats, Scientz, JY92-IIN). The solution was then centrifuged at 3,000×g for 10 min at 4°C. The supernatant was transferred to 4 mL of buffer B [40 mM Tris–HCl, pH 7.4, 600 mM NaCl, 4% (v/v) Triton X-114] and stirred at 4°C for 1 h to extract the membrane proteins. After being centrifuged at 10,000×g for 10 min at 4°C, the supernatant was incubated at 30°C for 3 min, and then centrifuged at 2,000×g for 20 min at 25°C. Upper (aqueous) phase was removed and lower (detergent) phase was collected. Proteins in the detergent phase were recovered by overnight precipitation at −40°C with 10 volumes of acetone and subsequent centrifugation at 10,000×g for 30 min at 4°C. The protein pellet was stored at −80°C.
To prepare rabbit anti-serum against RA bacterin (inactivated whole-cell bacteria), two New Zealand White rabbits (female, 2–3 kg) were injected subcutaneously with 0.5 mL of 109 formaldehyde inactivated WJ4 whole cells (0.04% v/v at 4°C for 24 h) in Montanide ISA 50V (SEPPIC, France) adjuvant three times at 4-week intervals. Blood samples were collected 2 weeks after the third injection. The antiserum was isolated and tested for enzyme-linked immunosorbent assay (ELISA) titers as described (Silva et al. 1998). A titer above 1:10,000 qualifies the antiserum to be used in Western blotting and bactericidal assay.
Two-dimensional gel electrophoresis and Western blotting
Two-dimensional gel electrophoresis (2-DE) was performed in duplicates for Coomassie Brilliant Blue G-250 staining and Western blotting procedures. The acetone-precipitated proteins were suspended in 0.5 mL of re-swelling buffer [8 M urea, 2% CHAPS, 2% v/v immobilized pH gradient (IPG) buffer, pH 4–7, 20 mM DTT, and 0.002% bromophenol blue] and quantified by 2-D Quant Kit (GE Healthcare, Piscataway, NJ). Isoelectric focusing (IEF) in the first dimension was performed with IPG strips (11 cm, pH 4–7; Bio-Rad, Hercules, CA, USA) and PROTEAN IEF cell (Bio-Rad). After rehydration of 350 μg proteins at 50 V for 12 h, IEF was conducted at a constant temperature of 20°C at 250 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, 8,000 V for 4 h, and finally reached a total of 40,000 V h. Then, the IPG strips were equilibrated for 15 min in equilibration buffer (50 mM Tris–HCl, pH 8.8, 6 M urea, 30% v/v glycerol, 2% w/v sodium dodecyl sulfate (SDS), 0.002% bromophenol blue, and 10 mg/mL dithiothreitol), followed by 15 min in above equilibration buffer containing 25 mg/mL of iodoacetamide. SDS polyacrylamide gel electrophoresis (SDS-PAGE) in the second dimension was carried out using 12% polyacrylamide gels. After 2-DE, one gel was stained with Coomassie Brilliant Blue G-250 (0.1% CBB G-250, 34% methanol, 17% ammonium sulfate, and 3% orthophosphoric acid) and scanned with Image Scanner (Amersham Pharmacia Biotech) to visualize 2-DE profiles, while the other gel was used for Western blotting with rabbit antiserum against RA bacterin to identify immunogenic proteins.
For Western blotting, the proteins on the gel were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) by semi-dry blotting with blotting buffer (100 mM borate, 20% methanol, pH 9.0) for 2 h at 1 mA/cm2. The membrane was blocked with blocking buffer [5% skim milk, 0.05% Tween-20 in phosphate-buffered saline (PBS)] for 2 h at room temperature, washed with PBST, and then incubated with rabbit anti-serum against RA bacterin (1:1,000) for 1 h followed by horseradish peroxidase (HRP)-labeled anti-rabbit IgG (1:5,000, GE Healthcare) for 1 h. The membrane was washed with PBST, equilibrated in 50 mM Tris–HCl buffer (pH 7.4) and then developed with 3,3′-diaminobenzidine (DAB, Sigma, St. Louis, MO, USA) until optimum color development was observed. All samples were performed in triplicate.
MALDI-TOF-MS and database searches
The Coomassie-stained protein spots corresponding to the spots on the Western blotting membrane were manually excised from the gel and transferred to a V-bottom 96-well plate for tryptic in-gel digestion. After digestion, the peptide mixtures were suspended in matrix solution [0.5 g/L alpha-cyano-4-hydroxy-cinnamic acid in ethanol–acetone solution (2:1 in v/v)], spotted on the matrix-assisted laser desorption/ionization (MALDI) plate and then analyzed by a 4700 MALDI time of flight (MALDI-TOF) Proteomics Analyzer (Applied Biosystems). Mass accuracy for peptide mass fingerprint (PMF) was searched in MASCOT (http://www.matrixscience.com). Theoretical isoelectric point (pI) and molecular weight (MW) for confident spots were calculated using the ExpaSy Protparam tool (http://www.expasy.org/tools/). Protein with a score >75 was valued as significant (p < 0.05). Lower scoring proteins were either verified manually or rejected. NCBI (http://www.ncbi.nlm.nih.gov) database and Sanger database (http://www.sanger.ac.uk) searches were conducted to obtain the corresponding DNA sequences.
groEL amplification by genome walking
Chromosomal DNA from RA strain WJ4 was isolated using Wizard Genomic DNA Purification Kit according to the manufacturer’s protocol. Primers GroEF1 and GroER1 used to amplify the partial sequence of the gene were designed by DNASTAR software based on the homologous/consensus sequence of groEL in GenBank (NC_002663, NC_003155, NC_009838, NC_006155, NC_010698, and NC_000964). The PCR products were cloned into TA-cloning vectors and sequenced.
The whole gene sequence for WJ4 groEL was obtained by genome walking as described (Siebert et al. 1995) with modifications. Specifically, four gene-specific primers SP1, SP2, SP3, and SP4 were synthesized according to the known partial sequence of WJ4 groEL. Genome Walker Adaptor and adaptor primers WP1 and WP2 were designed as described with minor modifications (Siebert et al. 1995, Table 1). To prepare the adaptor-ligated DNA, five restriction digestions were set up, where 2.5 μg of isolated genomic DNA was digested overnight at 37°C in a 100 μL reaction volume containing 1 U/μL of either EcoRV, PvuII, ScaI, StuI, or SmaI. Each batch of digested genomic DNA was cleaned up with GeneJETTM PCR Purification Kit according to the manufacturer’s protocol (Fermentas), then ligated separately to the Genome Walker Adaptor by mixing 1 μg digested genomic DNA, 5 μM adaptor and 0.5 U/μL T4 DNA ligase in a total volume of 20 μL and then incubating overnight at 16°C. Ligation only worked for EcoRV digested genomic DNA so the EcoRV digested and adaptor ligated DNA library was used as the Genome Walker library for the subsequent PCR-based walking reactions. The 5′ and 3′ walking procedures were performed separately using the Genome Walker Library as the template for primary PCR reactions and the primary PCR products as the templates for the secondary PCR reactions. For 5′ walking, gene-specific primer SP1 and adaptor primer WP1 were used for the primary reaction; nested gene-specific primer SP2 and adaptor primer WP2 were used for the secondary reaction. For 3′ walking, gene-specific primer SP3 and adaptor primer WP1 were used for the primary reaction; nested gene-specific primer SP4 and adaptor primer WP2 were used for the secondary reaction. The cycle parameters for both primary and secondary reactions were as the following: (94°C–25 s, 72°C–3 min) × 7 cycles, (94°C–25 s, 67°C–3 min) × 32 cycles, 72°C–7 min. The secondary PCR products for 5′ and 3′ walking were sequenced.
Expression and purification of WJ4 GroEL
The groEL gene in WJ4 was amplified by PCR using oligonucleotides GroEL-F and GroEL-R (Table 1). The resulting PCR product was digested with endonucleases SalI and BamHI and cloned into SalI/BamHI-digested expression vector pGEX-6P-1 to construct pGEX-GroEL. E. coli BL21 was transformed with pGEX-GroEL and the expression of recombinant GroEL (rGroEL) was induced by 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG.). rGroEL expression was analysed by SDS-PAGE and Western blotting as described above. rGroEL expressed in E. coli BL21 were further purified using GST Bind Purification Kit (Merck, San Diego, CA, USA) according to manufacturer’s protocol.
Immunization assay
Immunogenicity of rGroEL was examined in both ducklings and rabbits. Forty-eight 10-day-old Cherry Valley ducklings were divided into six groups of eight. The ducklings in groups 1, 3, and 5 received two subcutaneous injections on the neck of saline in Montanide ISA VG (SEPPIC) adjuvant as negative controls; other ducklings in group 2, 4, and 6 received two subcutaneous injections on the neck of 250 μg purified rGroEL in Montanide ISA VG (SEPPIC) adjuvant. The two injections were given 2 weeks apart, and all ducklings were bled at 10 days after the second injection to test the ELISA titers.
Two New Zealand rabbits were injected twice subcutaneously with 1 mg purified GroEL in Montanide ISA 50V (SEPPIC) adjuvant for each time. The two injections were given 2 weeks apart. Blood samples were collected before injection and 10 days after the second injection, and anti-sera were isolated to test the ELISA titers and bactericidal activities.
Care and maintenance of all animals were in accordance with the Institutional Animal Care and Use Committee guidelines set by Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Serum anti-GroEL ELISA units were detected using purified rGroEL as a coating antigen. Specifically, 96-well ELISA plates were overnight coated with 1 μg/well sample of rGroEL in 100 μL bicarbonate buffer (pH 9.6). The plates were overnight blocked with 5% skim milk power (w/v) in PBS at 4°C. The duckling or rabbit anti-sera were diluted in 2-fold steps from 1:500 to 1:16,000 with PBST; 100 μL of each diluted anti-sera was added to the well for 2 h incubation at 37°C. Two wells per plate with no addition of anti-serum were used as the negative control. After extensive washing with PBST, the plates were incubated with 100 μL/well of HRP-conjugated goat anti-duck IgG antisera (1:5,000, Sigma) or HRP-conjugated anti-rabbit IgG antisera (1:4,000, Sigma) for 1 h at 37°C for respective detection of duckling or rabbit anti-sera. The reaction were developed with 100 μL of tetramethyl benzidine substrate solution (Sigma) for 20 min of incubation at 37°C and terminated with 50 μL 2 M H2SO4, the OD450 of each well was read on an ELISA reader (Biotek, USA). The highest dilution of the sera with their OD450 value of >2.1 times than negative control wells were valued as the ELISA titers. The geometric mean ELISA titer of the negative control sera from ducklings in groups 1, 3, and 5 or rabbits before injection was assigned 1 as the ELISA unit. The ELISA units of the anti-sera were calculated as geometric mean ELISA titer of anti-sera/geometric mean ELISA titer of negative control sera. All the samples were performed in triplicate.
Bactericidal assay
Rabbit pre-serum (before injection) and rabbit anti-sera against rGroEL or RA bacterin were inactivated at 56°C for 30 min and tested for the bactericidal activity against RA strains with different serotypes using a microbactericidal assay as described (Yu and Gu 2005) with modifications. Specifically, each well of a 96-well plate contained 50 μL of rabbit anti-sera or pre-serum; then, 30 μL of bacterial suspension (104 CFU/mL in Dulbecco’s PBS containing calcium, magnesium, and 0.1% gelatin, DPBSG) and 20 μL of rabbit complement sera (1:5 in DPBSG; Sigma) were added. After incubation at 37°C for 60 min, 50 μL of the mixture was plated onto TSA plates. The plates were incubated at 37°C overnight with 5% CO2, and the colonies were counted. A total of six RA strains were tested, and experiments for each strain were performed in triplicates. The killing percentage was obtained by calculating the geometric mean value of [1 − (CFU from anti-serum/CFU from pre-serum)] × 100 from the three trials.
Animal challenge experiment
RA serotype 1 strain WJ4, serotype 2 strain Th4, and serotype 10 strain YXb-2 were used for the animal challenge experiment. Before challenge, the median lethal dose (LD50) for each strain was measured as described (Hu et al. 2010). On day 14 after two immunizations of 250 μg purified rGroEL in Montanide ISA VG (SEPPIC) adjuvant, ducklings of groups 1 and 2 were challenged with strains WJ4, group 3s and 4 were challenged with strains Th4, and groups 5 and 6 were challenged with strains YXb-2, respectively, at a challenge dose of 3 LD50 to evaluate the protection of rGroEL against RA challenge.
PCR amplification and sequence analysis for groEL in the 34 additional RA strains
Chromosomal DNAs from the 34 RA strains other than WJ4 were prepared using Wizard Genomic DNA Purification Kit. The groEL sequences were amplified by PCR with primers GroEL-F and GroEL-R according to the following cycle parameters: 94°C–4 min, (94°C–40 s, 50°C–30 s, 72°C–1 min) × 30 cycles, 72°C–10 min. PCR products were analyzed by electrophoresis on 1% agarose gels and groEL from nine RA strains (8785, D743, D26220, HXb-2, JY-1, NJ-2, NJ-3, P2123, and R20) were sequenced. Sequence homology among the ten groELs (including WJ4 groEL) was analyzed with DNASTAR software.
Statistical analysis
Antibody levels are expressed as the geometric mean ELISA units of n independent observations ± standard deviation. Significance was determined with the two-tailed independent Student’s t test, and p < 0.05 were considered significant. Protection from animal challenge is expressed as the protection rate. Significance was determined with chi-square test with SPSS 16.0, and p < 0.05 were considered significant, otherwise, the p values were indicated.
Results
Identification of the immunogenic outer membrane proteins in WJ4
2-DE was performed to separate the proteins in the sample. Coomassie-staining showed dozens of spots on the gel (Fig. 1a). Western blotting with rabbit anti-serum against RA bacterin showed four DAB-stained immunogenic proteins on the PVDF membrane (Fig. 1b). Locations corresponding to these four spots were marked as 1, 2, 3, and 4 on the Coomassie-stained gel (Fig. 1a).
Coomassie-stained 2-DE gel profile for outer membrane proteins of RA strain WJ4 and Western blotting. a Coomassie-stained 2-DE gel profile. Spots marked with 1, 2, 3, and 4 represent the spots that were immunoblotted with rabbit anti-serum against RA on Western blotting membrane. b Western blotting profile. Spots 1, 2, 3, and 4 were immunoblotted
Coomassie-stained spots 1–4 were excised and analyzed by MALDI-TOF mass spectrometry (MALDI-TOF-MS). According to the PMF and MASCOT search results, spots 1–3 were identified as OmpA precursor (protein score = 411), OmpA (protein score = 285), and putative GroEL (protein score = 126), respectively. OmpA precursor and OmpA appeared as distinct spots on the 2-DE gels because OmpA precursor contains a signal polypeptide with different pI and MW from those of mature OmpA. Theoretical pI and MW for WJ4 GroEL were calculated to be 4.93 and 57 kDa, respectively. Spot 4 has not to be characterized yet since its protein score was below 75.
Expression, purification and immunization of WJ4 GroEL
A 968-bp DNA fragment was amplified from WJ4 genomic DNA with primers GroEF1 and GroER1. Genome walking in attempt to obtain the full-length groEL, rendered a 1.8-kb fragment upstream and a 0.9-kb fragment downstream of the partial WJ4 groEL sequence. With the newly obtained 5′ and 3′, WJ4 groEL sequence was aligned with other bacterial groEL sequences obtained from GenBank using DNASTAR software. The full-length WJ4 groEL gene was determined to be 1,629 bp long, encoding 543 amino acids. The nucleotide sequence has been deposited in the GenBank database under accession number GU060633. With basic local alignment search tool analysis, the deduced RA GroEL amino acid sequence exhibits 91% or 90% amino acid identity with the GroEL in Chryseobacterium gleum (ATCC 35910) or Flavobacteriaceae bacterium (3519-10), respectively, and >80% amino acid identity with other GroEL sequences, such as Flavobacteria bacterium (MS024-3 C), Gramella forsetii (KT0803), Kordia algicida (OT-1), Robiginitalea biformata (HTCC2501), etc. It indicated that GroEL protein of RA had extensive homology with that of other bacteria.
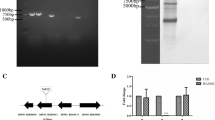
The recombinant expression vector pGEX-GroEL was transformed into E. coli BL21. rGroEL expression was analyzed by SDS-PAGE and Western blotting. After SDS-PAGE, the gel was stained with Coomassie Brilliant Blue G-250. E. coli BL21 transformed with pGEX-GroEL showed a band of rGroEL with glutathione sulfatransferase (Fig. 2a, lane 1), while E. coli BL21 did not (Fig. 2a, lane 2). Western blotting showed blot band at the corresponding location with the expression of rGroEL for transformed E. coli BL21 (Fig. 2b, lane 1). The results revealed the rGroEL was successfully expressed in E. coli BL21. Furthermore, rGroEL was purified from 2,000 mL of log-phase cultured E. coli BL21 transformed with pGEX-GroEL. A total of 36 mg of rGroEL was obtained for immunization assay. After two injections, anti-GroEL antibodies in immunized ducklings or rabbits were 53- or 160-fold higher, respectively, than in their non-immunized counterparts (Table 2).
The expression of rGroEL in E. coli BL21. a Coomassie-stained SDS-PAGE profiles. b Western blotting profile. Lane M Prestained protein ladder (Fermentas, SM6071). Lanes 1 E. coli BL21 transformed with expression vector pGEX-GroEL. Lanes 2 E. coli BL21. Lane 3 Purified rGroEL. Arrows denote the expression of rGroEL
Bactericidal activity of rabbit anti-sera
The anti-sera against GroEL of RA showed 30.0–57.3% cross-killing of RA strains containing different serotypes, whereas the anti-serum against bacterin of RA showed 0.2–63.6% killing of these strains, exhibited much greater variations to different serotype strains (Table 3).
Animal experiment
The LD50 of strains WJ4, Th4, and YXb-2 were 5.43 × 108, 6.35 × 107, and 3.54 × 106, respectively. After challenge, the death of duckling was recorded daily for a period of 7 days. The result showed that the ducklings immunized with rGroEL were 50%, 37.5%, and 37.5% protected from the challenge with WJ4, Th4, and YXb-2, respectively (Table 4).
Homology analysis of groEL in different serotype of RA strains
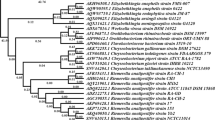
PCR using WJ4 primers successfully amplified groEL from all 34 RA strains tested, a first indication that the gene is highly conserved. groEL from nine RA strains with different serotypes were chosen for sequencing. DNA sequence analysis showed that the sequence identity of groEL between WJ4 and the nine additional strains was more than 97.5% despite their different serotypes (Fig. 3). It is inferred that such high conservation also applies to groEL from the other 25 strains, so they were not sequenced.
Percent identity and divergence analysis of RA groELs by DNASTAR software. The sequences of groEL from 10 RA strains with different serotypes were analyzed for percent identity and divergence. The homology of the sequences was >97.5%. The sequences of groEL from 8785, D743, JY-1, NJ-2, NJ-3, P2123, and WJ4 showed 100% identities
Discussion
In the present study, we performed immunoproteomics to search for more immunogenic proteins of RA for potential vaccine candidates and identified a 57-kDa GroEL as a novel one, which provided 37.5–50% protection for immunized ducklings from the challenge of both homologous and heterogeolous serotypes of RA strains.
GroEL belongs to the chaperonin family of molecular chaperones; it is highly conserved among prokaryotic and eukaryotic cells. The exact function of GroEL has not been precisely determined. Some GroEL proteins have been studied for their chaperone function in protein processing and assembly or for their cooperation with other chaperones (particularly GroES) (Tang et al. 2006; Wang et al. 1998). Other GroEL proteins are thought to protect intracellular pathogens against the hostile environment of host phagocytic cells (Fields et al. 1986). GroEL proteins are also reported to be potent immunogens in a number of infections. Using immunoproteomics, GroEL of Neospora caninum has been identified to be immunogenic protein (Shin et al. 2004). Immunization with GroEL of Bacillus anthracis provides 100% protection against Bacillus anthracis infection in BALB/c mice (Sinha and Bhatnagar 2010). Mice immunized with Salmonella GroEL produced a significant increased antibody titers, passive immunization with anti-GroEL sera protected 50% mice against lethal Salmonella infection (Khan et al. 2009). GroEL has also shown immunogenicity and protection against Streptococcus pneumoniae, Burkholderia pseudomallei, and Francisella tularensis infections (Hartley et al. 2004; Woo et al. 2001). There is no report concerning RA GroEL protein and its encoding gene yet, although large numbers of other bacteria GroELs have been reported (Zeilstra-Ryalls et al. 1991). In order to investigate the immunological characteristics of RA GroEL, immunization assay, bactericidal assay and animal challenge experiment were performed. As shown in “Results,” purified RA rGroEL elicited significant higher level of antibodies against rGroEL in both ducklings and rabbits. Importantly, bactericidal assay showed that the rabbit anti-serum against rGroEL killed 30.0–57.3% of RA strains with different serotypes 1, 2, and 10, whereas the rabbit anti-serum against RA bacterin showed a huge variation in killing of strains with different serotypes. Here, killing percentages ranged from 0.2% to 63.6%. This indicates that the anti-rGroEL serum kills a broad range of RA serotypes in a more consistent fashion than anti-bacterin serum that is more serotype-dependent. Animal challenge experiment with RA serotype 1, 2, or 10 strains showed that the immunized ducklings were 50%, 37.5%, and 37.5% protected, respectively, the similar broad range protection as that in bactericidal assay. Although the protection rate is not as high as that of inactivated bacterin (>80% protection, data not shown), rGroEL of RA did show some extent protection and cross-protection against the challenge of RA strains presenting different serotypes. To our knowledge, this is the first report regarding the cross-protection of RA immunogenic protein, which might contribute to the GroEL is the most conserved proteins known. The further study may lead to making RA GroEL a potential vaccine candidate against RA infections.
References
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422:198–207
Cooper GL (1989) Pasteurella anatipestifer infections in California turkey flocks: circumstantial evidence of a mosquito vector. Avian Dis 33:809–815
Cullen PA, Cordwell SJ, Bulach DM, Haake DA, Adler B (2002) Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect Immun 70:2311–2318
Fields PI, Swanson RV, Haidaris CG, Heffron F (1986) Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA 83:5189–5193
Han X, Aslanian A, Yates JR (2008) Mass spectrometry for proteomics. Curr Opin Chem Biol 12:483–490
Hartley MG, Green M, Choules G, Rogers D, Rees DG, Newstead S, Sjostedt A, Titball RW (2004) Protection afforded by heat shock protein 60 from Francisella tularensis is due to copurified lipopolysaccharide. Infect Immun 72:4109–4113
Helfer DH, Helmboldt CF (1977) Pasteurella anatipestifer infection in turkeys. Avian Dis 21:712–715
Hu Q, Han X, Zhou X, Ding S, Ding C, Yu S (2010) Characterization of biofilm formation by Riemerella anatipestifer. Vet Microbiol 144:429–436
Huang B, Subramaniam S, Frey J, Loh H, Tan HM, Fernandez CJ, Kwang J, Chua KL (2002) Vaccination of ducks with recombinant outer membrane protein (OmpA) and a 41 kDa partial protein (P45N’) of Riemerella anatipestifer. Vet Microbiol 84:219–230
Kardos G, Nagy J, Antal M, Bistyak A, Tenk M, Kiss I (2007) Development of a novel PCR assay specific for Riemerella anatipestifer. Lett Appl Microbiol 44:145–148
Khan MN, Shukla D, Bansal A, Mustoori S, Ilavazhagan G (2009) Immunogenicity and protective efficacy of GroEL (hsp60) of Streptococcus pneumoniae against lethal infection in mice. FEMS Immunol Med Microbiol 56:56–62
Pathanasophon P, Phuektes P, Tanticharoenyos T, Narongsak W, Sawada T (2002) A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol 31:267–270
Sandhu T (1979) Immunization of White Pekin ducklings against Pasteurella anatipestifer infection. Avian Dis 23:662–669
Shin YS, Lee EG, Shin GW, Kim YR, Lee EY, Kim JH, Jang H, Gershwin LJ, Kim DY, Kim YH, Kim GS, Suh MD, Jung TS (2004) Identification of antigenic proteins from Neospora caninum recognized by bovine immunoglobulins M, E, A and G using immunoproteomics. Proteomics 4:3600–3609
Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23:1087–1088
Silva I, Dangolla A, Allen J (1998) Seroepidemiology of rinderpest in bovids in Sri Lanka using the enzyme linked immunosorbent assay (ELISA) technique. Prev Vet Med 37:69–75
Sinha K, Bhatnagar R (2010) GroEL provides protection against Bacillus anthracis infection in BALB/c mice. Mol Immunol 48:264–271
Sinha S, Kosalai K, Arora S, Namane A, Sharma P, Gaikwad AN, Brodin P, Cole ST (2005) Immunogenic membrane-associated proteins of Mycobacterium tuberculosis revealed by proteomics. Microbiology 151:2411–2419
Subramaniam S, Huang B, Loh H, Kwang J, Tan HM, Chua KL, Frey J (2000) Characterization of a predominant immunogenic outer membrane protein of Riemerella anatipestifer. Clin Diagn Lab Immunol 7:168–174
Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M (2006) Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125:903–914
Wang JD, Michelitsch MD, Weissman JS (1998) GroEL–GroES-mediated protein folding requires an intact central cavity. Proc Natl Acad Sci USA 95:12163–12168
Woo PC, Leung PK, Wong SS, Ho PL, Yuen KY (2001) GroEL encodes a highly antigenic protein in Burkholderia pseudomallei. Clin Diagn Lab Immunol 8:832–836
Yu S, Gu XX (2005) Synthesis and characterization of lipooligosaccharide-based conjugate vaccines for serotype B Moraxella catarrhalis. Infect Immun 73:2790–2796
Zeilstra-Ryalls J, Fayet O, Georgopoulos C (1991) The universally conserved GroE (Hsp60) chaperonins. Annu Rev Microbiol 45:301–325
Zhang W, Lu CP (2007) Immunoproteomics of extracellular proteins of Chinese virulent strains of Streptococcus suis type 2. Proteomics 7:4468–4476
Acknowledgments
We thank Dr. Guoqiang Zhu from Yangzhou University and Dr. Zhizhong Cui from Shangdong Agriculture University for providing RA serotype reference strains. This work was funded by the National Natural Science Foundation of China (31072161), Shanghai Key Project on Agricultural Development through Science and Technology (2009HNG5-3), and National Basic Fund for Research Institutes, which is supported by the Chinese Academy of Agricultural Sciences (2008JB16).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Han, X., Hu, Q., Ding, S. et al. Identification and immunological characteristics of chaperonin GroEL in Riemerella anatipestifer . Appl Microbiol Biotechnol 93, 1197–1205 (2012). https://doi.org/10.1007/s00253-011-3635-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3635-2