Abstract
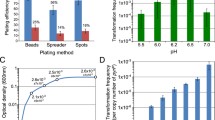
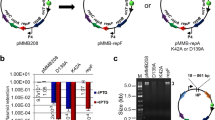
This paper reports the in vivo expression of the synthetic transposase gene tnp(a) from a hyperactive Tn5 tnp gene mutant in Streptomyces coelicolor. Using the synthetic tnp(a) gene adapted for Streptomyces codon usage, we showed random insertion of the transposon into the Streptomycetes genome. The insertion frequency for the hyperactive Tn5 derivative is 98% of transformed S. coelicolor cells. The random transposition has been confirmed by the recovery of ~1.1% of auxotrophs. The Tn5 insertions are stably inherited in the absence of apramycin selection. The transposon contains an apramycin resistance selection marker and an R6Kγ origin of replication for transposon rescue. We identified the transposon insertion loci by random sequencing of 14 rescue plasmids. The majority of insertions (12 of 14) were mapped to putative open-reading frames on the S. coelicolor chromosome. These included two new regulatory genes affecting S. coelicolor growth and actinorhodin biosynthesis.



Similar content being viewed by others
References
Anne J, van Mellaert L (1993) Streptomyces lividans as a host for heterologous protein production. FEMS Microbiol Letters 114(2):121–128
Ashour J, Hondalus MK (2003) Phenotypic mutants of the intracellular actinomycete Rhodococcus equi created by in vivo Himar1 transposon mutagenesis. J Bacteriol 185:2644–2652
Baltz RH, McHenney MA, Cantwell CA, Queener SW, Solenberg PJ (1997) Applications of transposition mutagenesis in antibiotic producing streptomycetes. Antonie van Leuwenhoek 71:179–187
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O´Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors fort the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116(1):43–49
Bishop A, Fielding S, Dyson P, Herron P (2004) Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaption and antibiotic production. Genome Res 14(5):893–900
Calderone T, Stevens RD, Oas TG (1996) High-level misincorporation oflysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J Mol Biol 262:407–412
Delic V, Hopwood DA, Friend EJ (1970) Mutagenesis by N-methyl-N’-nitro-N-nitrosoguanidine (NTG) in Streptomyces coelicolor. Mutat Res 9(2):167–182
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Fedoryshyn M, Petzke L, Welle E, Bechthold A, Luzhetskyy A (2008) Marker removal from actinomycetes genome using FLP recombinase. Gene 419:43–47
Flärdh K, Buttner M (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49
Herron P, Hughes G, Chandra G, Fielding S, Dyson P (2004) Transposon Express, a software application to report the identity of insertions obtained by comprehensive transposon mutagenesis of sequenced genomes: analysis of the preference for in vitroTn5 transposition into GC-rich DNA. Nucleic Acids Res 32:e113
Gehring AM, Nodwell JR, Beverley SM, Losick R (2000) Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. PNAS 97:9642–9647
Goryshin I, Jendrisak J, Hoffman L, Meis R, Reznikoff WS (2000) Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat Biotechnol 18:97–100
Grabher C, Wittbrodt J (2008) Recent advances in meganuclease-and transposon-mediated transgenesis of medaka and zebrafish. Methods Mol Biol 461:521–539
Guenes G, Smith B, Dyson P (1999) Genetic instability associated with insertion of IS6100 into one end of the Streptomyces lividans chromosome. Microbiology 145:2203–2208
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. PNAS 100:1541–1546
Ikeda H, Takada Y, Pang CH, Tanaka H, Omura S (1993) Transposon mutagenesis by Tn4560 and applications with avermectin-producing Streptomyces avermitilis. J Bacteriol 175(7):2077–2082
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21(5):526–531
Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. PNAS 100:14339–14344
Kane JF (1995) Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol 6:494–500
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Strepromyces genetics. John Innes Foundation, Norwich
Lyell NL, Dunn KA, Bose JL, Vescovi SL, Stabb E (2008) Effective mutagenesis of Vibrio fischeri using hyperactive mini-Tn5 derivatives. Appl Environ Microbiol 74:7059–7063
Luzhetskyy A, Fedoryshyn M, Gromyko O, Ostash B, Rebets Y, Bechthold A, Fedorenko V (2006) IncP plasmids are most effective in mediating conjugation between Escherichia coli and streptomycetes. Genetika 42:595–601
Matsushima P, Baltz RH (1987) recA gene of Escherichia coli complements defects in DNA repair and mutagenesis in Streptomyces fradiae JS6 (mcr-6). J Bacteriol 169(10):4834–4836
Mering C, Jensen L, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork, P (2007) STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35. doi:https://doi.org/10.1093/nar/gkl825
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25(4):447–453
Plasterk RH, Izsvák Z, Ivics Z (1999) Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Gen 15(8):326–332
Reznikoff WS (1993) The Tn5 transposon. Annu Rev Microbiol 47:945–963
Reznikoff WS (2003) Tn5 as a model for understanding DNA transposition. Genome Res 47(5):1199–1206
Reznikoff WS, Steininger-White MM, Metzler JD (2006) US patent 7083980—Tn5 transposase mutants and the use thereof.
Rholl D, Trunck L, Schweizer HP (2008) In vivo Himar1 transposon mutagenesis of Burkhoderia pseudomallei. Appl Environ Microbiol 74:7529–7535
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sandmann A, Frank B, Müller R (2008) A transposon-based strategy to scale up myxothiazol production in myxcobacterial cell factories. J Biotechnol 135(3):255–261
Schultz JE (2009) Structural and biochemical aspects of tandem GAF domains. Handb Exp Pharmacol 191:93–109
Solenberg PJ, Baltz RH (1991) Transposition of Tn5096 and other IS493 derivatives in Streptomyces griseofuscus. J Bacteriol 173:1096–1104
Weaden J, Dyson P (1998) Transposon mutagenesis with IS6100 in the avermectin-producer Streptomyces avermitilis. Microbiology 144:1963–1970
Wildenbrant E, Kao C (2007) Introduction of the foreign transposon Tn4560 in Streptomyces coelicolor leads to genetic instability near the native insertion sequence IS1649. J Bacteriol 189:9108–9116
Volff JN, Altenbuchner J (1997) High frequency transposition of the Tn5 derivative Tn5493 in Streptomyces lividans. Gene 194:81–86
Acknowledgment
We are very grateful to Prof. A. Bechthold for critical discussions during the work as well as for the financial support. The authors also wish to thank Dr. B. Gust for providing plasmid vector pIJ773. Elisabeth Welle is acknowledged for screening of the auxotrophic mutants. Dr. Annette Erb and Stephen Weber are acknowledged for reading and correcting this paper with accuracy.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at https://doi.org/10.1007/s00253-010-2511-9
Rights and permissions
About this article
Cite this article
Petzke, L., Luzhetskyy, A. In vivo Tn5-based transposon mutagenesis of Streptomycetes. Appl Microbiol Biotechnol 83, 979–986 (2009). https://doi.org/10.1007/s00253-009-2047-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2047-z