Abstract
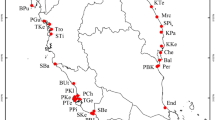
Bacterial communities associated with plant propagules remain understudied, despite the opportunities that propagules represent as dispersal vectors for bacteria to new sites. These communities may be the product of a combination of environmental influence and inheritance from parent to offspring. The relative role of these mechanisms could have significant implications for our understanding of plant–microbe interactions. We studied the correlates of microbiome community similarities across an invasion front of red mangroves (Rhizophora mangle L.) in Florida, where the species is expanding northward. We collected georeferenced propagule samples from 110 individuals of red mangroves across 11 populations in Florida and used 16S rRNA gene (iTag) sequencing to describe their bacterial communities. We found no core community of bacterial amplicon sequence variants (ASVs) across the Florida range of red mangroves, though there were some ASVs shared among individuals within most populations. Populations differed significantly as measured by Bray–Curtis dissimilarity, but not Unifrac distance. We generated data from 6 microsatellite loci from 60 individuals across 9 of the 11 populations. Geographic distance was correlated with beta diversity, but genetic distance was not. We conclude that red mangrove propagule bacterial communities are likely influenced more by local environmental acquisition than by inheritance.




Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study will be available in the NCBI SRA Database repository under submission numbers PRJNA844062 and PRJNA842720 upon acceptance of publication. [https://www.ncbi.nlm.nih.gov/sra/PRJNA844062, https://www.ncbi.nlm.nih.gov/sra/PRJNA842720].
References
Berg G, Rybakova D, Grube M, Köberl M (2016) The plant microbiome explored: implications for experimental botany. J Exp Bot 67:995–1002. https://doi.org/10.1093/jxb/erv466
Rodríguez CE, Antonielli L, Mitter B et al (2020) Heritability and functional importance of the setaria viridis bacterial seed microbiome. Phytobiomes J 4:40–52. https://doi.org/10.1094/PBIOMES-04-19-0023-R
Hacquard S (2016) Disentangling the factors shaping microbiota composition across the plant holobiont. New Phytol 209(2):454–457
Zilber-Rosenberg I, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. https://doi.org/10.1111/j.1574-6976.2008.00123.x
Tannenbaum I, Kaur J, Mann R et al (2020) Profiling the lolium perenne microbiome: from seed to seed. Phytobiomes J 4:281–289. https://doi.org/10.1094/PBIOMES-03-20-0026-R
Compant S, Clément C, Sessitsch A (2010) Soil biology & biochemistry plant growth-promoting bacteria in the rhizo- and endosphere of plants : their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42(5):669–678. https://doi.org/10.1016/j.soilbio.2009.11.024
Vokou D, Vareli K, Zarali E et al (2012) Exploring biodiversity in the bacterial community of the Mediterranean phyllosphere and its relationship with airborne bacteria. Microb Ecol 64:714–724. https://doi.org/10.1007/s00248-012-0053-7
Ul-Hasan S, Bowers RM, Figueroa-Montiel A et al (2019) Community ecology across bacteria, archaea and microbial eukaryotes in the sediment and seawater of coastal Puerto Nuevo, Baja California. PLoS One 14(2):e0212355
Cope-Selby N, Cookson A, Squance M et al (2017) Endophytic bacteria in Miscanthus seed: implications for germination, vertical inheritance of endophytes, plant evolution and breeding. GCB Bioenergy 9:57–77. https://doi.org/10.1111/gcbb.12364
Johnston-Monje D, Raizada MN (2011) Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6:e20396. https://doi.org/10.1371/journal.pone.0020396
Puente ME, Li CY, Bashan Y (2009) Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ Exp Bot 66:402–408. https://doi.org/10.1016/j.envexpbot.2009.04.007
Puente ME, Li CY, Bashan Y (2009) Rock-degrading endophytic bacteria in cacti. Environ Exp Bot 66:389–401. https://doi.org/10.1016/j.envexpbot.2009.04.010
Soldan R, Mapelli F, Crotti E et al (2019) Bacterial endophytes of mangrove propagules elicit early establishment of the natural host and promote growth of cereal crops under salt stress. Microbiol Res 223–225:33–43. https://doi.org/10.1016/j.micres.2019.03.008
Shade A, McManus PS, Handelsman J (2013) Unexpected diversity during community succession in the apple flower microbiome. MBio 4:e00602-12. https://doi.org/10.1128/mBio.00602-12
Singh P, Santoni S, This P, Péros J-PP (2018) Genotype-environment interaction shapes the microbial assemblage in grapevine’s phyllosphere and carposphere: an NGS approach. Microorganisms 6:96. https://doi.org/10.3390/microorganisms6040096
Drexler JZ (2001) Maximum longevities of Rhizophora apiculata and R. mucronata propagules. Pacific Sci 55:17–22. https://doi.org/10.1353/psc.2001.0004
Scherer B (2021) Patterns of bacterial community variation across anatomical, geographical, and genetic distance in Florida mangroves. Dissertation, Florida State University
Duke NC, Ball MC, Ellison JC et al (2017) Factors influencing biodiversity and distributional gradients in mangroves. Biogeogr Lett 7:27–47
Cavanaugh KC, Kellner JR, Forde AJ et al (2014) Poleward expansion of mangroves is a threshold response to decreased frequency of extreme cold events. Proc Natl Acad Sci U S A 111:723–727. https://doi.org/10.1073/pnas.1315800111
Koenig CC, Coleman FC, Eklund A-M et al (2007) Mangroves as essential nursery habitat for Goliath grouper (Epinephelus Itajara). Bull Mar Sci 80(3):567–585
Norton SL, Wiley TR, Carlson JK et al (2012) Designating critical habitat for juvenile endangered smalltooth sawfish in the United States. Mar Coast Fish 4:473–480. https://doi.org/10.1080/19425120.2012.676606
Morrissey JF, Gruber SH (1993) Habitat selection by juvenile lemon sharks, Negaprion brevirostris. Environ Biol Fishes 38:311–319. https://doi.org/10.1007/BF00007524
Hodel RGJ, De Souza Cortez MB, Soltis PS, Soltis DE (2016) Comparative phylogeography of black mangroves (Avicennia germinans) and red mangroves (Rhizophora mangle) in Florida: testing the maritime discontinuity in coastal plants. Am J Bot 103:730–739. https://doi.org/10.3732/ajb.1500260
Kennedy JP, Garavelli L, Truelove NK et al (2017) Contrasting genetic effects of red mangrove (Rhizophora mangle L.) range expansion along West and East Florida. J Biogeogr 44:335–347. https://doi.org/10.1111/jbi.12813
Kennedy JP, Craig H, Jara-Cavieres A et al (2020) Multiplex microsatellite PCR panels for the neotropical red mangrove, Rhizophora mangle: combining efforts towards a cost-effective and modifiable tool to better inform conservation and management. Conserv Genet Resour 12:503–513. https://doi.org/10.1007/s12686-020-01138-8
Moye ZD, Das CR, Tokman JI et al (2020) Treatment of fresh produce with a Salmonella-targeted bacteriophage cocktail is compatible with chlorine or peracetic acid and more consistently preserves the microbial community on produce. J Food Saf 40:e12763. https://doi.org/10.1111/jfs.12763
Trudeau S, Thibodeau A, Côté JC et al (2020) Contribution of the broiler breeders’ fecal microbiota to the establishment of the eggshell microbiota. Front Microbiol 11:1–14. https://doi.org/10.3389/fmicb.2020.00666
Herlemann DPR, Labrenz M, Jürgens K et al (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. https://doi.org/10.1038/ismej.2011.41
Illumina (2013) 16S metagenomic sequencing library. https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html. Accessed 10 Jan 2019
Boylen E, Rideout J, Dillon M et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Geneious (2021) Analyzing microsatellite traces tutorial. https://www.geneious.com/tutorials/microsatellites/. Accessed 10 Jan 2019
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Yilmaz P, Parfrey LW, Yarza P et al (2014) The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucl Acids Res 42:D643–D648. https://doi.org/10.1093/nar/gkt1209
Muthiah S, Corrada Bravo H (2020) Wrench: wrench normalization for sparse count data. R. Package version 1.8.0. https://github.com/HCBravoLab/Wrench
Chen J (2018) GUniFrac: generalized UniFrac distances. R package version 1:1
Oksanen J (2013) vegan: community ecology package. R package version 2.5-7. https://CRAN.R-project.org/package=vegan
Anderson MJ (2017) Permutational multivariate analysis of variance (PERMANOVA). Wiley statsref: Statistics Reference Online. Wiley, pp 1–15
Bray JR, Curtis JT (1957) An ordination of upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349
Griffiths SM, Antwis RE, Lenzi L et al (2019) Host genetics and geography influence microbiome composition in the sponge Ircinia campana. J Anim Ecol 88:1684–1695. https://doi.org/10.1111/1365-2656.13065
Parker KE, Ward JO, Eggleston EM et al (2020) Characterization of a thermally tolerant Orbicella faveolata reef in Abaco, The Bahamas. Coral Reefs 39:675–685. https://doi.org/10.1007/s00338-020-01948-0
Suárez-Moo P, Lamelas A, Garcia-Bautista I et al (2020) Characterization of sediment microbial communities at two sites with low hydrocarbon pollution in the southeast Gulf of Mexico. PeerJ 8:1–23. https://doi.org/10.7717/peerj.10339
Redford A, Bowers R, Knight R et al (2010) Variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893. https://doi.org/10.1111/j.1462-2920.2010.02258.x
Nelson EB (2017) The seed microbiome: origins, interactions, and impacts. Plant Soil 422:1–28. https://doi.org/10.1007/s11104-017-3289-7
Turner TR, James EK, Poole PS et al (2013) The plant microbiome. Genome Biol 14:209. https://doi.org/10.1186/gb-2013-14-6-209
Hodel RGJ, Chen S, Payton AC et al (2017) Adding loci improves phylogeographic resolution in red mangroves despite increased missing data: comparing microsatellites and RAD-Seq and investigating loci filtering. Sci Rep 7:1–14. https://doi.org/10.1038/s41598-017-16810-7
Cregger MA, Veach AM, Yang ZK et al (2018) The Populus holobiont: dissecting the effects of plant niches and genotype on the microbiome. Microbiome 6:1–14. https://doi.org/10.1186/s40168-018-0413-8
Acknowledgements
The authors thank the following individuals and organizations: Katelyn Desorcy-Scherer, Olivia Mason, Scott Steppan, Emily Lemmon, Sophie McCoy, Peter Beerli, Fritz Pichardo, Natasza Fontaine, Prashant Singh, Amber Brown, Steven Miller, Catalina Cuellar-Gempeler, Jackson Powell, Kevin Olson, Natali Ramirez-Bullon, Cheston Peterson, Brian Moe, Catelyn Snyder, the Apalachicola National Estuarine Reserve, the Florida Department of Environmental Protection, the FSU Coastal and Marine Lab, the FSU Department of Biological Sciences, the FSU Graduate School, FNPS, and FNPS Magnolia Chapter.
Funding
This work was supported by the Florida Native Plant Society (FNPS); FNPS Magnolia Chapter; Florida State University (FSU) Department of Biological Sciences; and the FSU Graduate School.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Brendan Scherer. The first draft of the manuscript was written by Brendan Scherer, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Scherer, B.P., Mast, A. Red Mangrove Propagule Bacterial Communities Vary With Geographic, But Not Genetic Distance. Microb Ecol 86, 1010–1022 (2023). https://doi.org/10.1007/s00248-022-02147-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02147-w