Abstract
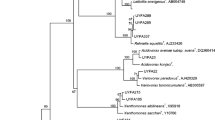
This study used a multiphasic approach, characterized by the simultaneous use of culture-dependent and culture-independent methods, to investigate endophytic bacterial communities in strawberry (Fragaria ananassa) fruit. A total of 92 bacterial endophytes were isolated and initially grouped by their repetitive extragenic palindromic (rep)-PCR banding pattern and biochemical features. Phylogenetic analysis of the 16S rRNA gene sequences of 45 representatives showed that the isolates belonged to the species Bacillus subtilis (eight isolates), Bacillus sp. (seven isolates), Enterobacter sp. (seven isolates), Enterobacter ludwigii (six isolates), Lactobacillus plantarum (six isolates), Pseudomonas sp. (five isolates), Pantoea punctata (three isolates), and Curtobacterium citreum (three isolates). Nucleic acids were extracted from the strawberry fruit and subjected to 16S rRNA gene directed polymerase chain reaction denaturing gradient gel electrophoresis (16S rRNA PCR-DGGE). The species B. subtilis, Enterobacter sp., and Pseudomonas sp. were detected both by isolation and DGGE. The DGGE fingerprints of total bacterial DNA did not exhibit bands corresponding to several of the representative species isolated in the extinction dilution (L. plantarum, C. citreum, and P. punctata). In contrast, bands in the DGGE profile that were identified as relatives of Arthrobacter sp. and one uncultivable Erythrobacter sp. were not recovered by cultivation techniques. After isolation, the nitrogen fixation ability and the in vitro production of indole-3-acetic acid (IAA) equivalents and siderophores were evaluated. A high percentage of isolates were found to possess the ability to produce siderophores and IAA equivalents; however, only a few isolates belonging to the genera Pseudomonas and Enterobacter showed the ability to fix nitrogen. Plant growth promotion was evaluated under greenhouse conditions and revealed the ability of the Bacillus strains to enhance the number of leaves, shoot length, root dry weight, and shoot dry weight. The activity of the bacterial isolate identified as B. subtilis NA-108 exerted the greatest influence on strawberry growth and showed a 42.8% increase in number of leaves, 15.26% for high shoot, 43.5% increase in root dry weight, and a 77% increase in shoot dry weight when compared with untreated controls.




Similar content being viewed by others
References
Andreote FD, Carneiro RT, Salles JF, Marcon J, Labate CA, Azevedo JL, Araújo WL (2009) Culture-independent assessment of Alphaproteobacteria related to order Rhizobiales and the diversity of cultivated Methylobacterium in the rhizosphere and rhizoplane of transgenic eucalyptus. Microb Ecol 57:82–93
Andreote FD, Rossetto PB, Mendes R, Avila LA, Labate CA, Pizzirani-Kleiner AA, Azevedo JL, Araújo WL (2009) Bacterial community in the rhizosphere and rhizoplane of wild type and transgenic eucalyptus. World J Microbiol Biotechnol 25:1065–1073
Araújo WL, Marcon J, Maccheroni W Jr, van Elsas JD, van Vuurde JWL, Azevedo JL (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68:4906–4914
Berg G (2009) Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11–18
Chelius MK, Triplett EW (2001) The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol 41:252–263
Dias ACF, Costa FEC, Andreote FD, Lacava PT, Teixeira MA, Assumpção LC, Araújo WL, Azevedo JL, Melo IS (2009) Isolation of micropropagated strawberry endophytic bactéria and assessment of their potential for plant growth promotion. World J Microbiol Biotechnol 25:189–195
Esitken A, Yildiz S, Ercisli S, Donmez MF, Turan M, Gunes A (2007) Effects of plant growth promoting bacteria (PGPB) on yield, growth and nutrient contents of organically grown strawberry. Appl Microbiol Biotechnol 76:1145–1152
Forchetti G, Masciarelli O, Alemano S, Alvarez D, Abdala G (2007) Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl Microbiol Biotechnol 76:1145–1152
Funk G, Hutson RA, Bernard KA, Pfyffer GE, Wauters G, Collins MD (2006) Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J Clin Microbiol 34:2356–2363
Fürnkranz M, Müller H, Berg G (2009) Characterization of plant growth promoting bacteria from crops in Bolivia. J Plant Dis Protec 116(4):149–155
Garbeva P, Overbeek LSV, Van Vuurde JWL, Van Elsas JD (2001) Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microb Ecol 41:369–383
Garbeva P, Postma J, vanVeen JA, vanElsas JD (2006) Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ Microbiol 8:233–246
Gevers D, Huys G, Swigs J (2001) Applicability of rep-PCR fingerprinting for identification of species. FEMS Microbiol Lett 205:31–36
Glick BR (1995) The enhancement of plant growth by free-living bacteria. Can J Microbiol 41:109–117
Gordon SA, Weber RP (1951) Colorimetric estimation of indole acetic acid. Plant Physiol 26:192–195
Hallman J, Quadt-Hallman A, Rodriuez-Kabana R, Kloepper JW (1998) Interactions between Meloidogyne incognita and endophytic bacteria in cotton and cucumber. Soil Biol Biochem 30:925–937
Holt JG, Krieg NR, Sneath PHA, Stanley JT, Williams ST (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams & Wilkins, Baltimore, p 787
Idris R, Trifonova R, Puschenreiter M, Wenzel WW, Sessitsch A (2004) Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl Environ Microbiol 70:2667–2677
Jayne-Williams DJ (1976) The application of miniaturized methods for the characterization of various organisms isolated from the animal gut. J Appl Bacteriol 40:189–200
Kloepper JW (1991) Development of in vivo assays for prescreening antagonists of Rhizoctonia solani on cotton. Phythopathol 81:1006–1013
Krimm U, Abanda-Nkpwatt D, Schwab W, Schreiber L (2005) Epiphytic microorganisms on strawberry plants (Fragaria ananassa cv. Elsanta): identification of bacterial isolates and analysis of their interaction with leaf surfaces. FEMS Microbiol Ecol 53:483–492
Kuklinsky-Sobral J, Araújo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AL, Azevedo JL (2004) Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ Microbiol 12:1244–1251
Leach AW, Mumford JD (2008) Pesticide environmental accounting: a method for assessing the external costs of individual pesticide applications. Environ Pollut 151:139–147
Li C-H, Zhao M-W, Tang C-M, Li S-P (2010) Population dynamics and identification of endophytic bacteria antagonistic toward plant-pathogenic fungi in cotton root. Microb Ecol 59:344–356
Liu Z, Sinclair J (1993) Colonization of soybean roots by Bacillus megaterium B153-2-2. Soil Biol Biochem 25:849–855
Loaces I, Ferrando L, Scavino AF (2010) Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb Ecol 61:606–618
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Magalhães K, Pereira MA, Nicolau A, Dragone G, Domingues L, Teixeira JA, de Almeida Silva JB, Schwan RF (2010) Production of fermented cheese whey-based beverage using kefir grains as starter culture: Evaluation of morphological and microbial variations. Biores Tech 101:8843–8850
Magnusson JK, Strom S, Roos J, Sjogren J, Schnurer J (2003) Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol Lett 219:129–135
Monteiro JM, Vollú RE, Coelho MRR, Alviano CS, Blank AF, Seldin L (2009) Comparison of the bacterial community and characterization of plant growth-promoting rhizobacteria from different genotypes of Chrysopogon zizanioides (L.) Roberty (Vetiver) rhizospheres. J Microbiol 4:363–370
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electroforesis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nadkarni MA, Martin FE, Hunter N, Jacques NA (2009) Methods for optimizing DNA extraction before quantifying oral bacterial numbers by real-time PCR. FEMS Microbiol Lett 296:45–51
Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH (2007) The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and cultureindependent methods. Int J Food Microbiol 114:168–186
Noel TC, Cheng C, Yost CK, Pharis RP, Hynes MF (1996) Rhizobium leguminosarum as a plant-growth rhizobacterium: direct growth promotion of canola and lettuce. Can J Microbiol 42:279–283
Ovreås L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373
Pedraza RO, Motok J, Salazar SM, Ragout AL, Mentel MI, Tortora ML, Guerrero-Molina MF, Winik BC, Díaz-Ricci JC (2009) Growth-promotion of strawberry plants inoculated with Azospirillum brasilense. World J Microbiol Biotechnol 26:265–272
Pereira GVM, Ramos CL, Galvão C, Dias ES, Schwan RF (2010) Use of specific PCR primers to identify three important industrial species of Saccharomyces genus: Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces pastorianus. Lett Appl Microbiol 5:131–137
Roesch LFW, Quadros PD, Camargo FAO, Triplett EW (2007) Screening of diazotrophic bacteria Azopirillum spp. for nitrogen fixation and auxin production in multiple field sites in southern Brazil. World J Microbiol Biotechnol 23:1377–1383
Rosenblueth M, Martínez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant-Microb Interact 19:827–837
Sagaram US, De Angelis KM, Trivedi P, Andersen GL, Lu SE, Wang N (2009) Bacterial diversity analysis of Huanglongbing pathogen-infected citrus, using PhyloChip arrays and 16S rRNA gene clone library sequencing. Appl Environ Microbiol 75:1566–1574
Scheirlinck I, der Meulen RV, Vancanneyt M, De Vuyst L, Vandamme P, Huys G (2008) Taxonomic structure and stability of the bacterial community in Belgian sourdough ecosystems as assessed by culture and population fingerprinting. Appl Environ Microbiol 74:2414–2423
Sharma A, Johri BN (2003) Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS9 in maize (Zea mays L.) under iron limiting conditions. Microbiol Res 158:243–248
Shrestha A, Toyota K, Okazaki M, Suga Y, Quevedo MA, Loreto A, Mariscal AA (2007) Enhancement of nitrogen-fixing activity of Enterobacteriaceae strains isolated from Sago Palm (Metroxylon sagu) by microbial interaction with non-nitrogen fixers. Microbes Environ 22:59–70
Sun L, Qiu F, Zhang X, Dai X, Dong X, Song W (2008) Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb Ecol 55:415–424
Torriani S, Felis G, Dellaglio F (2001) Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl Environ Microbiol 67:3450–3454
Trivedi P, Duan Y, Wang N (2010) Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl Environ Microbiol 76:3427–3436
Trivedi P, Spann T, Wang N (2011) Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb Ecol. doi:10.1007/s00248-011-9822-y
Tulipani S, Mezzetti B, Battino M (2009) Impact of strawberries on human health: insight into marginally discussed bioactive compounds for the Mediterranean diet. Publ health nutrit 12:1656–1662
Ulrich K, Ulrich A, Ewald D (2008) Diversity of endophytic bacterial communities in poplar grown under field conditions. FEMS Microbiol Ecol 63:169–180
Wang X, Haruta S, Wang P, Ishii M, Igarashi Y, Cui Z (2006) Diversity stable enrichment culture which is useful for silage inoculant and its succession in alfalfa silage. Fed Euro Microbiol Soc 57:106–115
Weyant RS, Moss CW, Weaver RE, Frollis DG, Jordan JG, Cook EC, Daneshvar MI (1995) Identification of unusual pathogenic G negative aerobic and anaerobic bacteria. The Williams and Wilkins Company, Baltimore, pp 318–341
Zaidi S, Usmani S, Singh BR, Musarrat J (2006) Significance of Bacillus subtilis strain SJ-101 as a bioinoculant for concurrent plant growth promotion and nickel accumulation in Brassica juncea. Chemosphere 64:991–997
Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, Ishimaru C, Arunakumari A, Barletta RG, Vidaver AK (2002) Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208
Zoetendal EG, Akkermans ADL, de Vos WM (1998) Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable host-specific communities of active bacteria. Appl Environ Microbiol 64:3854–3859
Acknowledgments
The authors acknowledge Horto Florestal e Plantas Medicinais (UFLA) for providing the strawberry samples for use in this study and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à pesquisa do Estado de Minas Gerais (FAPEMIG) for financial support and Dr. Disney Ribeiro Dias for collaboration in manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Melo Pereira, G.V., Magalhães, K.T., Lorenzetii, E.R. et al. A Multiphasic Approach for the Identification of Endophytic Bacterial in Strawberry Fruit and their Potential for Plant Growth Promotion. Microb Ecol 63, 405–417 (2012). https://doi.org/10.1007/s00248-011-9919-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9919-3