Abstract
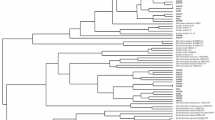
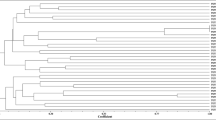
The aim of this work is to describe the diversity and phylogeny of rhizobial bacteria associated to nodules of Ononis tridentata L. in different geographical regions of Spain. Twenty-two bacterial isolates were characterized using several molecular techniques (16S amplified ribosomal deoxyribonucleic acid restriction analysis, fingerprinting, and sequencing) and phylogenies were inferred from their 16S and nodC gene sequences. Phylogenetically, the isolates grouped with the genera Rhizobium, Mesorhizobium, Phylobacterium, and Bosea. The nodC gene, essential for nodulation, was detected for the first time in isolates close to the genera Bosea and Phyllobacterium. The bacteria isolated showed a high diversity at the genus, species, and strain level regardless of the geographical origin of the host plant. This is the first report describing bacteria associated to nodules of O. tridentata. This shrub legume is highly prized for the revegetation of gypsum soils in semiarid Mediterranean areas. Our molecular description of bacteria associated to this legume improves the current understanding of the ecology of this plant species. Our findings have implications for formulating suitable bacterial inocula to recover gypsum ecosystems.




Similar content being viewed by others
References
Aguilar OM, López MV, Donato M, Morón B, Soria-Diaz ME, Mateos C, Gil-Serrano A, Sousa C, Megías M (2006) Phylogeny and nodulation signal molecule of rhizobial populations able to nodulate common beans other than the predominant species Rhizobium etli present in soils from the northwest of Argentina. Soil Biol Biochem 38:573–586
Council of the European Community (1992) Directive 92/43 of the Council of the European Community on the conservation of habitats and wild fauna and flora. European Community, Brussels, Belgium
Barret CF, Parker MA (2006) Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp nodule bacteria on two Mimosa spp. in Costa Rica. Appl Environm Microbiol 72:1198–1206
Benhizia Y, Benhizia H, Benguedouar A, Muresu R, Giacomini A, Squartini A (2004) Gamma proteobacteria can nodulate legumes of the genus Hedysarum. Syst Appl Microbiol 27:462–468
Brockwell J, Bottomley PJ, Thies JE (1995) Manipulation of rhizobia microflora for improving legume productivity and soil fertility. Plant and Soil 174:143–180
Chen W-M, Moulin L, Bontemps C, Vandamme P, Béna G, Boivin-Masson C (2003) Legume symbiotic nitrogen fixation by β-Proteobacteria is widespread in nature. J Bacteriol 185:7266–7272
Dana ED, Mota JF (2006) Vegetation and soil recovery on gypsum outcrops in semi-arid Spain. J Arid Environm 65:444–459
Elliot GN, Chen W-M, Chou J-H, Wang H-C, Sheu S-Y, Perin L, Reis VM, Moulin L, Simon MF, Bontemps C, Sutherland JM, Bessi R, de Faria SM, Trinick MJ, Prescott AL, Sprent JI, James EK (2007) Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol 173:168–180
Garrity GM, Bell JA, Lilburn TG (2003) Taxonomic outline of Prokaryotes. In: Garrity GM (ed) Bergey’s manual of systematic bacteriology. 2nd edn. Springer, New York
Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, Bena G, Hannibal L, Fardoux J, Kojadinovic M, Vuillet L, Lajus A, Cruveiller S, Rouy Z, Mangenot S, Segurens B, Dossat C, Franck WL, Chang WS, Saunders E, Bruce D, Richardson P, Normand P, Dreyfus B, Pignol D, Stacey G, Emerich D, Vermeglio A, Medigue C, Sadowsky M (2007) Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316 (5829):1307–1312
González-Andrés J, Alegre J, Ceresuela JL (2005) The rhizobia nodulating shrubs for revegetation of arid lands: isolation of native strains and specificity of the plant–rhizobia interaction by cross inoculation tests. Arid Land Res Manag 19:307–326
Hall BG (2004) Phylogenetic trees made easy: A how-to manual, 2nd edn. Sinauer, Sunderland, MA
Hirsch CF, Sigmund JM (1995) Use of polymerase chain reaction (PCR) fingerprinting to differentiate bacteria for microbial products screening. J Indust Microbiol 15:85–93
Kalita M, Stepkowski T, Lotock B, Malek W (2006) Phylogeny of nodulation genes and symbiotic properties of Genista tinctoria bradyrhizobia. Arch Microbiol 186:87–97
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981–993
Lane DJ (1991) 16S/23S rRNA sequencing. In: Strackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Chischester, Wiley, pp 115–175
Liu J, Wang ET, Chen WX (2005) Diverse rhizobia associated with woody legumes Wisteria sinensis, Cercis racemosa and Amorpha fruticosa grown in the temperate zone of China. Syst Appl Microbiol 28:465–477
Lucas MM, Peart JL, Brewin NJ, Kannenberg EL (1996) Isolation of monoclonal antibodies reacting with the core component of lipopolysaccharide from Rhizobium leguminosarum strain 3841 and mutant derivatives. J Bacteriol 178:2727–2733
Mantelin S, Fisher-Le Saux M, Zakhia F, Béna G, Bonneau S, Jeder H, de Lajudie P, Cleyet-Marel JC (2006) Emended description of the genus Phyllobacterium and description of four novel species associated with plant roots: Phyllobacterium bourgognense sp. nov., Phyllobacterium ifriqiyense sp. nov., Phyllobacterium leguminum sp. nov. and Phyllobacterium brassiccearum sp. nov. Int J Syst Evol Microbiol 56:827–839
Moschetti G, Peluso AL, Protopapa A, Anastasio M, Pepe O, Defez R (2005) Use of nodulation pattern, stress tolerance, nodC gene amplification, RAPD-PCR and RFLP–16S rDNA analysis to discriminate genotypes of Rhizobium leguminosarum biovar viciae. Syst Appl Microbiol 28:619–631
Odee DW, Haukka K, McInroy SG, Sprent JI, Sutherland JM, Young JPW (2002) Genetic and symbiotic characterisation of rhizobia isolated from tree and herbaceous legumes grown in soils from ecologically diverse sites in Kenya. Soil Biol Biochem 34:801–811
Palacio S, Escudero A, Montserrat-Martí G, Maestro M, Milla R, Albert MJ (2007) Plants living on gypsum: beyond the specialist model. Ann Bot 99:333–343
Perret X, Staehelin C, Broughton WJ (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Res 64:180–201
Pugnaire FI, Lázaro R (2000) Seed bank and understory species composition in a semi-arid environment: the effect of shrub age and rainfall. Ann Bot 86:807–813
Rambaut A (1996) Se-Al: sequence alignment editor. Available at http://evolve.zoo.ox.ac.uk/
Rasolomampianina R, Bailly X, Fetiarison R, Bena G, Ramaroson L, Raherimandimby M, Moulin L, de Lajudie P, Dreyfus B, Avarre JC (2005) Nitrogen-fixing nodules from rose wood legume trees (Dalbergia spp.) endemic to Madagascar host seven different genera belonging to (α- and β-Proteobacteria). Mol Ecol 14:4135–4146
Requena N, Jiménez I, Toro M, Barea JM (1997) Interactions between plant-growth promoting rhizobacteria (PGPR), arbuscular mycorrhizal fungi and Rhizobium spp. in the rhizosphere of Anthyllis cytisoides, a model legume for revegetation in the Mediterranean semi-arid ecosystems. New Phytol 136:667–677
Requena N, Pérez-Solís E, Azcón-Aguilar C, Jeffries P, Barea JM (2001) Management of indigenous plant-microbe symbioses aids restoration of desertified ecosystems. Appl Environ Microbiol 67:495–498
Rodriguez-Echeverría S, Pérez-Fernández MA (2005) Potential use of Iberian shrubby legumes and rhizobia inoculation in revegetation projects under acidic soil conditions. Appl Soil Ecol 29:203–208
Romdhane SB, Nasr H, Samba-Mbaye R, Neyra M, Ghorbal MH, de Lajudie P (2006) Genetic diversity of Acacia tortilis ssp. raddiana rhizobia in Tunisia assessed by 16S and 16S-23S rDNA genes analysis. J Appl Microbiol 100:436–445
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Fritsch EF, Manniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Sarita S, Sharma PK, Priefer UB, Prell J (2005) Direct amplification of rhizobia nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol Ecol 54:1–11
Sawada H, Kuykendall LD, Young JM (2003) Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J Gen Appl Microbiol 49:155–179
Sneath PHA, Sokal RR (1973) Numerical taxonomy. WH Freeman, San Francisco
Storms V, Van den Vreken N, Gillis M, Vandamme P (2002) Evaluation of tRNA intergenic length polymorphism (tDNA-PCR) for the differentiation of the members of the Burkholderia cepacia complex. Syst Appl Microbiol 25:376–385
Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4. Sinauer, Sunderland, MA
Sy A, Giraud E, Jurad P, García N, Willems A, de Lajudie P, Prin Y, Neyra M, Gillis M, Bombin-Masson C, Dreyfus B (2001) Methylotropic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol 183:214–220
Thrall PH, Millsom DA, Jeavons AC, Waayers M, Harvey GR, Bagnall DJ, Brockwell J (2005) Seed inoculation with effective root-nodule bacteria enhances revegetation success. J Appl Ecol 42:740–751
Thompsom JD, Gibson TJ, Plewniak F, Jeanmoungin F, Higgins DG (1997) The CUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trujillo ME, Willems A, Abril A, Planchuelo AM, Rivas R, Ludeña D, Mateos PF, Martínez-Molina E, Velázquez E (2005) Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl Environ Microbiol 71:1318–1327
Ueda T, Suga Y, Yahiro N, Matsuguchi T (1995) Phylogeny of Sym plasmids of Rhizobia by PCR-based sequencing of a nodC segment. J Bacteriol 177:48–472
Valverde A, Velázquez E, Fernández-Santos F, Vizcaino N, Rivas R, Mateos PF, Martínez-Molina E, Igual JM, Willems A (2005) Phyllobacterium trifolii sp. nov., nodulating Trifolium and Lupinus in Spanish soils. Int J Syst Evol Micr 55:1985–1989
van Berkum P, Eardly BD (1998) Molecular evolutionary systematics of the Rhizobiaceae. In: Spaink HP, Kondorosi A, Hooykaas PJJ (eds) The Rhizobiaceae: molecular biology of model plant-associated bacteria. Kluwer, Dordrecht, pp 1–24
Vincent JM (1970) A manual for the practical study of root-nodules bacteria. IBP Handbook. Blackwell, Oxford, UK
Vinuesa P, Silva C, Lorite MJ, Izaguirre-Mayoral ML, Bedmar EJ, Martínez-Romero E (2005) Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst Appl Microbiol 28:702–716
Wang ET, Martínez-Romero E (2000) Sesbania herbacea–Rhizobium huautlense nodulation in flooded soils and comparative characterization of S. herbacea-nodulating Rhizobia in different environments. Microb Ecol 41:25–32
Weir BS, Turner SJ, Silvester WB, Park D-C, Young JM (2004) Unexpectedly diverse Mesorhizobium strains and Rhizobium leguminosarum nodulate native legume genera of New Zealand, while introduced legume weeds are nodulated by Bradyrhizobium species. Appl Environ Microbiol 70:5980–5987
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Young JP, Haukka KE (1996) Diversity and phylogeny of rhizobia. New Phytol 133:87–94
Young JM, Park DC, Weir BS (2004) Diversity of 16S rRNA sequences of Rhizobium spp. implications for species determinations. FEMS Microbiol Lett 238:125–131
Zakhia F, Jeder H, Domergue O, Willems A, Cleyet-Marel JC, Gillis M, Dreyfus B, de Lajudie P (2004) Characterisation of wild legume nodulating bacteria (LNB) in the infra-arid zone of Tunisia. Syst Appl Microbiol 27:380–395
Zakhia F, Jeder H, Willems A, Gillis M, Dreyfus B, de Lajudie P (2006) Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb Ecol 51:375–393
Zerhari K, Aurag J, Khbaya B, Kharchaf D, Filali-Maltouf A (2000) Phenotypic characteristics of rhizobia isolates nodulating Acacia species in the arid and Saharan regions of Morocco. Lett Appl Microbiol 30:351–357
Acknowledgments
The authors thank Dr. Noelle Amarger, Dr. Herman Spaink, and Dr. Encarna Velázquez for providing some of the reference strains. The authors thank Dr David Badía and Dr. Francisco Pugnaire for the localization of Ononis populations. We thank Monika Oggerin for assistance with phylogenetic analysis. This work was supported by grants GR/AMB/0735/2004 and S-0505/AMB/0321 from the Comunidad de Madrid (Spain). A. Rincón holds a I3P postdoctoral fellowship awarded by the Consejo Superior de Investigaciones Científicas (CSIC, Spain), and F. Arenal was awarded a postdoctoral grant by the Comunidad de Madrid.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Rincón and F. Arenal contributed equally to this work.
Rights and permissions
About this article
Cite this article
Rincón, A., Arenal, F., González, I. et al. Diversity of Rhizobial Bacteria Isolated from Nodules of the Gypsophyte Ononis tridentata L. Growing in Spanish Soils. Microb Ecol 56, 223–233 (2008). https://doi.org/10.1007/s00248-007-9339-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9339-6