Abstract
Lipidomics investigates the composition and function of lipids, typically employing blood or tissue samples as the primary study matrices. Hair has recently emerged as a potential complementary sample type to identify biomarkers in early disease stages and retrospectively document an individual’s metabolic status due to its long detection window of up to several months prior to the time of sampling. However, the limited coverage of lipid profiling presented in previous studies has hindered its exploitation. This study aimed to evaluate the lipid coverage of hair using an untargeted liquid chromatography-high-resolution mass spectrometry lipidomics platform. Two distinct three-step exhaustive extraction experiments were performed using a hair metabolomics one-phase extraction technique that has been recently optimized, and the two-phase Folch extraction method which is recognized as the gold standard for lipid extraction in biological matrices. The applied lipidomics workflow improved hair lipid coverage, as only 99 species could be annotated using the one-phase extraction method, while 297 lipid species across six categories were annotated with the Folch method. Several lipids in hair were reported for the first time, including N-acyl amino acids, diradylglycerols, and coenzyme Q10. The study suggests that hair lipids are not solely derived from de novo synthesis in hair, but are also incorporated from sebum and blood, making hair a valuable matrix for clinical, forensic, and dermatological research. The improved understanding of the lipid composition and analytical considerations for retrospective analysis offers valuable insights to contextualize untargeted hair lipidomic analysis and facilitate the use of hair in translational studies.
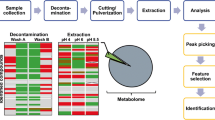
Graphical abstract





Similar content being viewed by others
Data availability
Raw datafiles are available through the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/) with the data set identifier MSV000091771.
References
Jang WJ, Choi JY, Park B, Seo JH, Seo YH, Lee SS, et al. Hair metabolomics in animal studies and clinical settings. Molecules. 2019;24(12):2195.
Robbins CR. Chemical and physical behavior of human hair. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. 130–147, 417 p.
Pragst F, Balikova MA. State of the art in hair analysis for detection of drug and alcohol abuse. Clin Chim Acta Int J Clin Chem. 2006;370(1–2):17–49.
Csuka DA, Csuka EA, Juhász MLW, Sharma AN, Mesinkovska NA. A systematic review on the lipid composition of human hair. Int J Dermatol. 2023;62(3):404–15.
Lyman DJ, Murray-Wijelath J. Fourier transform infrared attenuated total reflection analysis of human hair: comparison of hair from breast cancer patients with hair from healthy subjects. Appl Spectrosc. 2005;59(1):26–32.
Lyman DJ, Fay SG. The effect of breast cancer on the fourier transform infrared attenuated total reflection spectra of human hair. Ecancermedicalscience. 2014;8:405
Corino GL, French PW. Diagnosis of breast cancer by X-ray diffraction of hair. Int J Cancer. 2008;122(4):847–56.
Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4(7):594–610.
Yang L, Li M, Shan Y, Shen S, Bai Y, Liu H. Recent advances in lipidomics for disease research. J Sep Sci. 2016;39(1):38–50.
Han X. Lipidomics for studying metabolism. Nat Rev Endocrinol. 2016;12(11):668–79.
Ni Z, Wölk M, Jukes G, Mendivelso Espinosa K, Ahrends R, Aimo L, et al. Guiding the choice of informatics software and tools for lipidomics research applications. Nat Methods. 2023;20(2):193–204.
Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11(8):593–8.
Witting M, Böcker S. Current status of retention time prediction in metabolite identification. J Sep Sci. 2020;43(9–10):1746–54.
Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. TrAC Trends Anal Chem. 2014;61:192–206.
Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509.
Ulmer CZ, Jones CM, Yost RA, Garrett TJ, Bowden JA. Optimization of Folch, Bligh-Dyer, and Matyash sample-to-extraction solvent ratios for human plasma-based lipidomics studies. Anal Chim Acta. 2018;1037:351–7.
Eisenbeiss L, Steuer AE, Binz TM, Baumgartner MR, Kraemer T. (Un)targeted hair metabolomics: first considerations and systematic evaluation on the impact of sample preparation. Anal Bioanal Chem. 2019;411(17):3963–77.
Chang WCW, Wang PH, Chang CW, Chen YC, Liao PC. Extraction strategies for tackling complete hair metabolome using LC-HRMS-based analysis. Talanta. 2021;223(Pt 1): 121708.
Chen Y, Guo J, Xing S, Yu H, Huan T. Global-scale metabolomic profiling of human hair for simultaneous monitoring of endogenous metabolome, short- and long-term exposome. Front Chem. 2021;9:674265
da Silva KM, Iturrospe E, Heyrman J, Koelmel JP, Cuykx M, Vanhaecke T, et al. Optimization of a liquid chromatography-ion mobility-high resolution mass spectrometry platform for untargeted lipidomics and application to HepaRG cell extracts. Talanta. 2021;235: 122808.
Iturrospe E, Da Silva KM, Robeyns R, Van De Lavoir M, Boeckmans J, Vanhaecke T, et al. Metabolic signature of ethanol-induced hepatotoxicity in HepaRG cells by liquid chromatography-mass spectrometry-based untargeted metabolomics. J Proteome Res. 2022;21(4):1153–66.
Cooper GAA, Kronstrand R, Kintz P. Society of Hair Testing guidelines for drug testing in hair. Forensic Sci Int. 2012;218(1–3):20–4.
González-Domínguez R, González-Domínguez Á, Sayago A, Fernández-Recamales Á. Recommendations and best practices for standardizing the pre-analytical processing of blood and urine samples in metabolomics. Metabolites. 2020;10(6):229.
Smith L, Villaret-Cazadamont J, Claus SP, Canlet C, Guillou H, Cabaton NJ, et al. Important considerations for sample collection in metabolomics studies with a special focus on applications to liver functions. Metabolites. 2020;10(3):104.
Saini RK, Prasad P, Shang X, Keum YS. Advances in lipid extraction methods—a review. Int J Mol Sci. 2021;22(24):13643.
Iturrospe E, da Silva KM, van de Lavoir M, Robeyns R, Cuykx M, Vanhaecke T, et al. Mass spectrometry-based untargeted metabolomics and lipidomics platforms to analyze cell culture extracts. In New York, NY: Springer International Publishing; 2023. p. 189–206. (Methods in Molecular Biology; vol. 2571).
da Silva KM, Iturrospe E, van den Boom R, van de Lavoir M, Robeyns R, Vergauwen L, et al. Lipidomics profiling of zebrafish liver through untargeted liquid chromatography-high resolution mass spectrometry. J Sep Sci. 2022;45(15):2935–45.
Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–6.
Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, et al. A lipidome atlas in MS-DIAL 4. Nat Biotechnol. 2020;38(10):1159–63.
DeFelice BC, Mehta SS, Samra S, Čajka T, Wancewicz B, Fahrmann JF, et al. Mass Spectral Feature List Optimizer (MS-FLO): a tool to minimize false positive peak reports in untargeted liquid chromatography-mass spectroscopy (LC-MS) data processing. Anal Chem. 2017;89(6):3250–5.
Klåvus A, Kokla M, Noerman S, Koistinen VM, Tuomainen M, Zarei I, et al. “Notame”: workflow for non-targeted LC-MS metabolic profiling. Metabolites. 2020;10(4):1–35.
Koelmel JP, Kroeger NM, Ulmer CZ, Bowden JA, Patterson RE, Cochran JA, et al. LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics. 2017;18(1):331.
da Silva KM, van de Lavoir M, Robeyns R, Iturrospe E, Verheggen L, Covaci A, et al. Guidelines and considerations for building multidimensional libraries for untargeted MS-based metabolomics. Metabolomics. 2022;19(1):4.
Lange M, Angelidou G, Ni Z, Criscuolo A, Schiller J, Blüher M, et al. AdipoAtlas: a reference lipidome for human white adipose tissue. Cell Rep Med. 2021;2(10): 100407.
Liebisch G, Fahy E, Aoki J, Dennis EA, Durand T, Ejsing CS, et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J Lipid Res. 2020;61(12):1539–55.
Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Acevedo A, Durán C, Ciucci S, Gerl M, Cannistraci CV. LIPEA: lipid pathway enrichment analysis. bioRxiv. 2018;69:2749.
Bittremieux W, Wang M, Dorrestein PC. The critical role that spectral libraries play in capturing the metabolomics community knowledge. Metabolomics. 2022;18(12):94.
Mahieu NG, Patti GJ. Systems-level annotation of a metabolomics data set reduces 25 000 features to fewer than 1000 unique metabolites. Anal Chem. 2017;89(19):10397–406.
Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6(4):328–40.
Masukawa Y, Tsujimura H, Narita H. Liquid chromatography-mass spectrometry for comprehensive profiling of ceramide molecules in human hair. J Lipid Res. 2006;47(7):1559–71.
Bollinger JG, Rohan G, Sadilek M, Gelb MH. LC/ESI-MS/MS detection of FAs by charge reversal derivatization with more than four orders of magnitude improvement in sensitivity. J Lipid Res. 2013;54(12):3523–30.
Thomas MC, Altvater J, Gallagher TJ, Nette GW. Collision-induced dissociation of fatty acid [M – 2H + Na] – ions: charge-directed fragmentation and assignment of double bond position. J Am Soc Mass Spectrom. 2014;25(11):1917–26.
Lee WS, Oh TH, Chun SH, Jeon SY, Lee EY, Lee S, et al. Integral lipid in human hair follicle. J Investig Dermatol Symp Proc. 2005;10(3):234–7.
Lee WS. Integral hair lipid in human hair follicle. J Dermatol Sci. 2011;64(3):153–8.
Wertz PW, Downing DT. Integral lipids of mammalian hair. Comp Biochem Physiol B Comp Biochem. 1989;92(4):759–61.
Fu Z, Sinclair AJ. Novel pathway of metabolism of α-linolenic acid in the Guinea pig. Pediatr Res. 2000;47(3):414–7.
Wurst FM, Alexson S, Wolfersdorf M, Bechtel G, Forster S, Alling C, et al. Concentration of fatty acid ethyl esters in hair of alcoholics: comparison to other biological state markers and self reported-ethanol intake. Alcohol Alcohol. 2004;39(1):33–8.
Pragst F, Auwärter V, Kießling B, Dyes C. Wipe-test and patch-test for alcohol misuse based on the concentration ratio of fatty acid ethyl esters and squalene CFAEE/CSQ in skin surface lipids. Forensic Sci Int. 2004;143(2–3):77–86.
Eisenbeiss L, Binz TM, Baumgartner MR, Kraemer T, Steuer AE. Towards best practice in hair metabolomic studies: systematic investigation on the impact of hair length and color. Metabolites. 2020;10(10):1–12.
McCoin CS, Knotts TA, Adams SH. Acylcarnitines—old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol. 2015;11(10):617–25.
Bradshaw HB, Rimmerman N, Hu SSJ, Burstein S, Walker JM. Chapter 8 Novel endogenous N‐acyl glycines. In: Vitamins and hormones. Vitam Horm; 2009. p. 191–205.
Meckfessel MH, Brandt S. The structure, function, and importance of ceramides in skin and their use as therapeutic agents in skin-care products. J Am Acad Dermatol. 2014;71(1):177–84.
Nelson AB, Chow LS, Hughey CC, Crawford PA, Puchalska P. Artifactual FA dimers mimic FAHFA signals in untargeted metabolomics pipelines. J Lipid Res. 2022;63(5): 100201.
Masukawa Y, Tsujimura H. Highly sensitive determination of diverse ceramides in human hair using reversed-phase high-performance liquid chromatography–electrospray ionization mass spectrometry. Lipids. 2007;42(3):275–90.
Hussler G, Kaba G, Francois Am, Saint-Leger D. Isolation and identification of human hair ceramides. Int J Cosmet Sci. 1995;17(5):197–206.
Chaurasia B, Summers SA. Ceramides in metabolism: key lipotoxic players. Annu Rev Physiol. 2021;83(1):303–30.
Coderch L, López O, de la Maza A, Parra JL. Ceramides and skin function. Am J Clin Dermatol. 2003;4(2):107–29.
Singh EJ, Gershbein LL. Phospholipids of human hair lipids. J Chromatogr A. 1967;31(C):20–7.
Kimura A, Kanekura T, Saito Y, Sagawa K, Nosaka M, Kanzaki T, et al. Blood group A glycosphingolipid accumulation in the hair of patients with α-N-acetylgalactosaminidase deficiency. Life Sci. 2005;76(16):1817–24.
Luo Y, Zhang C, Ma L, Zhang Y, Liu Z, Chen L, et al. Measurement of 7-dehydrocholesterol and cholesterol in hair can be used in the diagnosis of Smith-Lemli-Opitz syndrome. J Lipid Res. 2022;63(6): 100228.
Leeder JD, Bishop DG, Jones LN. Internal lipids of wool fibers. Text Res J. 1983;53(7):402–7.
Picardo M, Ottaviani M, Camera E, Mastrofrancesco A. Sebaceous gland lipids. Dermato-Endocrinology. 2009;1(2):68–71.
Honda A, Yamashita K, Miyazaki H, Shirai M, Ikegami T, Xu G, et al. Highly sensitive analysis of sterol profiles in human serum by LC-ESI-MS/MS. J Lipid Res. 2008;49(9):2063–73.
Koskivuori J, Voutilainen R, Uusitalo L, Lehtonen M, Lakka T, Auriola S, et al. A quantitative ultra-performance liquid chromatography high-resolution mass spectrometry analysis of steroids from human scalp hair. J Pharm Biomed Anal. 2022;215: 114768.
Shackleton C, Pozo OJ, Marcos J. GC/MS in recent years has defined the normal and clinically disordered steroidome: will it soon be surpassed by LC/tandem MS in this role? J Endocr Soc. 2018;2(8):974–96.
Greff MJE, Levine JM, Abuzgaia AM, Elzagallaai AA, Rieder MJ, van Uum SHM. Hair cortisol analysis: an update on methodological considerations and clinical applications. Clin Biochem. 2019;63:1–9.
Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51(11):3299–305.
Passi S, De Pità O, Puddu P, Littarru GP. Lipophilic antioxidants in human sebum and aging. Free Radic Res. 2002;36(4):471–7.
Stewart ME. Sebaceous gland lipids. Semin Dermatol. 1992;11(2):100–5.
Akaza N, Akamatsu H, Numata S, Matsusue M, Mashima Y, Miyawaki M, et al. Fatty acid compositions of triglycerides and free fatty acids in sebum depend on amount of triglycerides, and do not differ in presence or absence of acne vulgaris. J Dermatol. 2014;41(12):1069–76.
Zákány N, Oláh A, Markovics A, Takács E, Aranyász A, Nicolussi S, et al. Endocannabinoid tone regulates human sebocyte biology. J Investig Dermatol. 2018;138(8):1699–706.
Westerberg R, Tvrdik P, Undén AB, Månsson JE, Norlén L, Jakobsson A, et al. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem. 2004;279(7):5621–9.
Lee L, Anthony DeBono C, Campagna DR, Young DC, Branch Moody D, Fleming MD. Loss of the acyl-CoA binding protein (Acbp) results in fatty acid metabolism abnormalities in mouse hair and skin. J Investig Dermatol. 2007;127(1):16–23.
van Hoogevest P, Fahr A. Phospholipids in cosmetic carriers. In: Nanocosmetics. Cham: Springer International Publishing; 2019. p. 95–140.
Acknowledgements
MvdL and EI acknowledge funding of the Research Scientific Foundation-Flanders (FWO)—project numbers 1120623N and 1161620N, respectively. KMdS was funded by the University of Antwerp (BOF DOCPRO 4-Antigoon ID 36893). In addition, RR acknowledges funding of the University of Antwerp (BOF-Antigoon ID 46315). The work was supported by the Exposome Centre of Excellence of the University of Antwerp (BOF grant, Antigoon database number 41222).
Author information
Authors and Affiliations
Contributions
MvdL: conceptualization, methodology, investigation, formal analysis, validation, visualization, writing—original draft, writing—review and editing. KMdS: methodology, investigation, writing—review and editing. EI and RR: methodology, writing—review and editing. ALNvN and AC: supervision, conceptualization, writing—review and editing, resources.
Corresponding authors
Ethics declarations
Ethical approval
Ethical approval for the use of hair samples was provided by the Medical Ethics Committee of the University Hospital Antwerp (reference number 1975).
Consent for publication
All authors critically read and approved the final submitted version of the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
van de Lavoir, M., da Silva, K.M., Iturrospe, E. et al. Untargeted hair lipidomics: comprehensive evaluation of the hair-specific lipid signature and considerations for retrospective analysis. Anal Bioanal Chem 415, 5589–5604 (2023). https://doi.org/10.1007/s00216-023-04851-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-023-04851-z