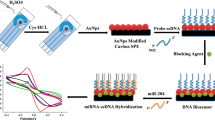
Abstract
Sensitive impedimetric detection of miR-222, a miRNA sequence found in many lung tumors, was investigated by using gold-nanostructured disposable carbon electrodes and enzyme-decorated liposomes. The proposed method was based on the immobilization of thiolated DNA capture probes onto gold-nanostructured carbon surfaces. Afterwards, the capture probes were allowed to hybridize to the target miRNAs. Finally, enzyme-decorated liposomes were used as labels to amplify the miRNA sensing, by their association with the probe–miRNA hybrids generated on the nanostructured transducer. By using this amplification route a limit of detection of 0.400 pM, a limit of quantification of 1.70 pM, and an assay range spanning three orders of magnitude (1.70–900 pM) were obtained (RSD % = 13). This limit of quantification was 20 times lower than that obtained using a simple enzyme conjugate for the detection. A comparison was also made with gold screen-printed transducers. In this case, a limit of quantification approximately 70 times lower was found by using the nanostructured transducers. Application of the optimized assay in serum samples was also demonstrated.

Alkaline Phosphatase-decorated liposomes and Au nanostructured screen-printed electrodes have been used for the impedimetric detection of miRNAs, via the bio-catalyzed precipitation of an insulating product onto the electrode surface.







Similar content being viewed by others
References
Suni II. Impedance methods for electrochemical sensors using nanomaterials. TrAC Trends Anal Chem. 2008;27:604–11. doi:10.1016/j.trac.2008.03.012.
Katz E, Willner I. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: routes to impedimetric immunosensors, DNA-sensors, and enzyme biosensors. Electroanalysis. 2003;15:913–47. doi:10.1002/elan.200390114.
Daniels JS, Pourmand N. Label-free impedance biosensors: opportunities and challenges. Electroanalysis. 2007;19:1239–57. doi:10.1002/elan.200603855.
Labuda J, Oliveira Brett AM, Evtugyn G, Fojta M, Mascini M, Ozsoz M, et al. Electrochemical nucleic acid-based biosensors: concepts, terms, and methodology (IUPAC Technical Report). Pure Appl Chem. 2010;82:1161. doi:10.1351/PAC-REP-09-08-16.
Edwards KA, Baeumner AJ. Optimization of DNA-tagged dye-encapsulating liposomes for lateral-flow assays based on sandwich hybridization. Anal Bioanal Chem. 2006;386:1335–43. doi:10.1007/s00216-006-0705-x.
Edwards KA, Curtis KL, Sailor JL, Baeumner AJ. Universal liposomes: preparation and usage for the detection of mRNA. Anal Bioanal Chem. 2008;391:1689–702. doi:10.1007/s00216-008-1992-1.
Chumbimuni-Torres KY, Wu J, Clawson C, Galik M, Walter A, Flechsig G-U, et al. Amplified potentiometric transduction of DNA hybridization using ion-loaded liposomes. Analyst. 2010;135:1618–23. doi:10.1039/C0AN00198H.
Kannuck RM, Bellama JM, Durst RA. Measurement of liposome-released ferrocyanide by a dual-function polymer modified electrode. Anal Chem. 1988;60:142–7. doi:10.1021/ac00153a009.
Alfonta L, Singh AK, Willner I. Liposomes labeled with biotin and horseradish peroxidase: a probe for the enhanced amplification of antigen − antibody or oligonucleotide − DNA sensing processes by the precipitation of an insoluble product on electrodes. Anal Chem. 2001;73:91–102. doi:10.1021/ac000819v.
Patolsky F, Lichtenstein A, Willner I. Electronic transduction of DNA sensing processes on surfaces: amplification of DNA detection and analysis of single-base mismatches by tagged liposomes. J Am Chem Soc. 2001;123:5194–205. doi:10.1021/ja0036256.
Patolsky F, Lichtenstein A, Willner I. Electrochemical transduction of liposome-amplified DNA sensing. Angew Chem Int Ed. 2000;39:940–3. doi:10.1002/(SICI)1521-3773(20000303)39:5<940::AID-ANIE940>3.0.CO;2-Y.
Palchetti I, Laschi S, Marrazza G, Mascini M. Electrochemical imaging of localized sandwich DNA hybridization using scanning electrochemical microscopy. Anal Chem. 2007;79:7206–13. doi:10.1021/ac070474h.
Soreta TR, Henry OYF, O'Sullivan CK. Electrode surface nanostructuring via nanoparticle electronucleation for signal enhancement in electrochemical genosensors. Biosens Bioelectron. 2011;26:3962–6. doi:10.1016/j.bios.2011.03.001.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi:10.1016/0092-8674(93)90529-Y.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci. 2002;99:15524–9. doi:10.1073/pnas.242606799.
Satoh J. Molecular network of microRNA targets in Alzheimer’s disease brains. Exp Neurol. 2012;235:436–46. doi:10.1016/j.expneurol.2011.09.003.
Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–92. doi:10.1111/j.1349-7006.2010.01650.x.
Palchetti I. Affinity biosensors for tumor-marker analysis. Bioanalysis. 2014;6:3417–35. doi:10.4155/bio.14.247.
Labib M, Berezovski MV. Electrochemical sensing of microRNAs: avenues and paradigms. Biosens Bioelectron. 2015;68:83–94. doi:10.1016/j.bios.2014.12.026.
Campuzano S, Pedrero M, Pingarrón JM. Electrochemical genosensors for the detection of cancer-related miRNAs. Anal Bioanal Chem. 2013;406:27–33. doi:10.1007/s00216-013-7459-z.
Teo AKL, Le Lim C, Gao Z. The development of electrochemical assays for microRNAs. Electrochim Acta. 2014;126:19–30. doi:10.1016/j.electacta.2013.06.113.
Bartosik M, Hrstka R, Palecek E, Vojtesek B. Adsorptive transfer stripping for quick electrochemical determination of microRNAs in total RNA samples. Electroanalysis. 2014;26:2558–62. doi:10.1002/elan.201400449.
Torrente-Rodríguez RM, Campuzano S, López-Hernández E, Granados R, Sánchez-Puelles JM, Pingarrón JM. Direct determination of miR-21 in total RNA extracted from breast cancer samples using magnetosensing platforms and the p19 viral protein as detector bioreceptor. Electroanalysis. 2014;26:2080–7. doi:10.1002/elan.201400317.
Erdem A, Congur G. Label-free voltammetric detection of microRNAs at multi-channel screen printed array of electrodes comparison to graphite sensors. Talanta. 2014;118:7–13. doi:10.1016/j.talanta.2013.09.041.
Tran HV, Piro B, Reisberg S, Duc HT, Pham MC. Antibodies directed to RNA/DNA hybrids: an electrochemical immunosensor for microRNAs detection using graphene-composite electrodes. Anal Chem. 2013;85:8469–74. doi:10.1021/ac402154z.
Kilic T, Nur Topkaya S, Ozsoz M. A new insight into electrochemical microRNA detection: a molecular caliper, p19 protein. Biosens Bioelectron. 2013;48:165–71. doi:10.1016/j.bios.2013.04.011.
Baydemir G, Bettazzi F, Palchetti I, Voccia D. Strategies for the development of an electrochemical bioassay for TNF-alpha detection by using a non-immunoglobulin bioreceptor. Talanta. 2016;151:141–7. doi:10.1016/j.talanta.2016.01.021.
Laschi S, Palchetti I, Marrazza G, Mascini M. Development of disposable low density screen-printed electrode arrays for simultaneous electrochemical measurements of the hybridisation reaction. J Electroanal Chem. 2006;593:211–8. doi:10.1016/j.jelechem.2006.04.015.
Bettazzi F, Hamid-Asl E, Esposito C, Quintavalle C, Formisano N, Laschi S, et al. Electrochemical detection of miRNA-222 by use of a magnetic bead-based bioassay. Anal Bioanal Chem. 2013;405:1025–34. doi:10.1007/s00216-012-6476-7.
Laschi S, Bulukin E, Palchetti I, Cristea C, Mascini M. Disposable electrodes modified with multi-wall carbon nanotubes for biosensor applications. ITBM-RBM. 2008;29:202–7. doi:10.1016/j.rbmret.2007.11.002.
Hoogvliet JC, Dijksma M, Kamp B, van Bennekom WP. Electrochemical pretreatment of polycrystalline gold electrodes to produce a reproducible surface roughness for self-assembly: a study in phosphate buffer pH 7.4. Anal Chem. 2000;72:2016–21. doi:10.1021/ac991215y.
Findlay JWA, Dillard RF. Appropriate calibration curve fitting in ligand binding assays. AAPS J. 2007;9:E260–7. doi:10.1208/aapsj0902029.
Giannetto M, Maiolini E, Ferri EN, Girotti S, Mori G, Careri M. Competitive amperometric immunosensor based on covalent linking of a protein conjugate to dendrimer-functionalised nanogold substrate for the determination of 2,4,6-trinitrotoluene. Anal Bioanal Chem. 2012;405:737–43. doi:10.1007/s00216-012-6137-x.
O’Mullane AP, Ippolito SJ, Sabri YM, Bansal V, Bhargava SK. Premonolayer oxidation of nanostructured gold: an important factor influencing electrocatalytic activity. Langmuir. 2009;25:3845–52. doi:10.1021/la8039016.
Hezard T, Fajerwerg K, Evrard D, Collière V, Behra P, Gros P. Gold nanoparticles electrodeposited on glassy carbon using cyclic voltammetry: application to Hg(II) trace analysis. J Electroanal Chem. 2012;664:46–52. doi:10.1016/j.jelechem.2011.10.014.
Komsiyska L, Staikov G. Electrocrystallization of Au nanoparticles on glassy carbon from HClO4 solution containing [AuCl4]−. Electrochim Acta. 2008;54:168–72. doi:10.1016/j.electacta.2008.08.013.
Zanardi C, Baldoli C, Licandro E, Terzi F, Seeber R. Development of a gold-nanostructured surface for amperometric genosensors. J Nanopart Res. 2012;14:1–12. doi:10.1007/s11051-012-1148-2.
Welch CM, Nekrassova O, Dai X, Hyde ME, Compton RG. Fabrication, characterisation and voltammetric studies of gold amalgam nanoparticle modified electrodes. ChemPhysChem. 2004;5:1405–10. doi:10.1002/cphc.200400263.
Bettazzi F, Lucarelli F, Palchetti I, Berti F, Marrazza G, Mascini M. Disposable electrochemical DNA-array for PCR amplified detection of hazelnut allergens in foodstuffs. Anal Chim Acta. 2008;614:93–102. doi:10.1016/j.aca.2008.03.027.
Acknowledgments
This work was supported by Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) in the framework of Progetti di ricerca di interesse nazionale (PRIN) 2012 (grant no. 20128ZZS2H) and Ente Cassa di Risparmio di Firenze project ID PED 8780 2014.0757A2202.0734.
EF and DB acknowledge CSGI (Consorzio per lo Sviluppo dei Sistemi a Grande Interfase) for partial financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest is declared.
Additional information
Published in the topical collection Chemical Sensing Systems with guest editors Maria Careri, Marco Giannetto, and Renato Seeber.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 387 kb)
Rights and permissions
About this article
Cite this article
Voccia, D., Bettazzi, F., Fratini, E. et al. Improving impedimetric nucleic acid detection by using enzyme-decorated liposomes and nanostructured screen-printed electrodes. Anal Bioanal Chem 408, 7271–7281 (2016). https://doi.org/10.1007/s00216-016-9593-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9593-x