Abstract
The objective of this work was to compare several models of body weight data from 90-day rodent feeding trials. Polynomial and nonlinear functions relating time and weight were examined as were the use of Toeplitz error covariance structures and random coefficients. The models were evaluated by fitting them to five publicly available datasets from rat feeding studies. Model performance was assessed in terms of their ability to capture the complexity of the growth patterns, validity of necessary assumptions, and information criteria scores. The results demonstrated the importance of selecting a curve function that effectively reflects the mean response. Toeplitz error covariance structures resulted in superior model fit, while failing to address deviations from model assumptions. Models using the Richards function and random coefficients were generally superior to the other models evaluated and dramatically improved upon linear models with complex error structures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Animal feeding studies are used to evaluate the safety of agricultural, pharmaceutical, industrial, and food chemicals. Additionally, if a compositional analysis of food or feed derived from a genetically modified (GM) crop suggests compositional differences from conventional crop varieties that could be potential health hazards, a 90-day feeding study in rodents has been recommended to provide additional information used in the comparative risk assessment (EFSA 2011a; ILSI Task Force 2004).
Internationally harmonized guidance on the design and conduct of 90-day toxicity studies has been developed by the Organisation for Economic Co-operation and Development ((OECD) 408 chemical testing guideline (2003)). The European Food Safety Authority (EFSA) requires certain adaptations of these methods for testing whole foods from GM crops (EFSA 2011b). This paper focuses on the statistical evaluation of data from studies following the EFSA guidance.
Traditionally, fixed effect linear models have been applied to compare treatment groups separately week-by-week, as described by Schmidt et al. (2016). As this approach considers each week separately, the chance of at least one false-positive result is inflated relative to comparing final weights among treatments. This approach also has reduced power to detect treatment effects that are small but consistent across time. Schmidt et al. (2016) proposed the use of linear mixed models with weight increasing linearly with time and a complex error correlation structure. This method improved interpretability by providing a single parameter to describe the response of weight over time. However, the linear relationship between weight and time prevents this model from estimating the curvature apparent in visual inspections of raw data curves from feeding trials. Polynomial or nonlinear curves of moderate complexity might be sufficient to fully capture the relationships. Kuhi et al. (2010) provide an extensive review of mathematical functions for modeling growth in poultry. Similar considerations should be made when evaluating options for rodent data.
The use of parametric structures to model patterns of correlation/covariance among error terms within an individual is well-established for models of repeated measurements (Littell et al. 2006, ch. 5). Such structures can capture the correlations between multiple measurements on the same rodent or cage, reflecting consistent effects of each individual rodent or cage. Additionally, error covariance structures can capture the tendency of measurements made close together in time to be more similar than those with greater separation.
An alternative to modeling covariance among repeated measures is the modeling of random coefficients in linear mixed models. Polynomial models with randomly distributed coefficients that follow parametric covariance structures are described by Vonesh and Chinchilli (1997) and Sjoblad et al. (1992). These models can also be adapted to non-polynomial relationships between the response and explanatory variables.
The objective of this work was to compare models for body weight data from 90-day rodent feeding trials. Polynomial and nonlinear functions were examined as were types of covariance structures. Performance of the models was compared in terms of empirical assessments of suitability for multiple trials.
Materials and methods
Motivating data
The motivating data we considered are from five rat feeding trials conducted as part of the European Commission funded GMO Risk Assessment and Communication of Evidence (GRACE) project. Detailed methods for the trials have been published previously (Schmidt et al. 2017; Zeljenková et al. 2014, 2016). Raw data and initial statistical analysis reports of all five trials can be found at the CADIMA website (https://www.cadima.info/index.php/area/publicAnimalFeedingTrials; search Project column for GRACE).
All five trials (denoted A, B, C, D, and E) included diets incorporating material from GM maize containing the same event at two different incorporation rates: 11% and 33%. We abbreviate these diets as GMO11 and GMO33. Diets made with material from a near-isogenic control of the GM maize were also included at the same incorporation rates in all five trials. Trials A, B, and C also included diets made with different varieties of conventional maize at the same incorporation rates.
In all trials, two rats of the same sex were housed in each cage and fed together, making cages the experimental units. Equal numbers of male and female rats were assigned to each diet treatment group. Each diet was fed to 8, 8, 10, 5, and 5 cages of male and female rats in trials A, B, C, D, and E, respectively. Rats used in the trials were selected to be relatively uniform in initial body weight, and were allocated to treatment groups such that the average weight among treatment groups was similar. Initiation of feeding on diet treatments and measurement of body weights was considered week 0 in modeling and graphing. Each rat was weighed weekly for 13 weeks (approximating 90 days) after feeding was begun. The duration of the 13th week was 5 days for trials A and B, and 6 days for trials D and E. All rats survived until the end of the studies, and there were no missing data. Although trial C was conducted for a full year, we only considered the first 90 days of weight data to match the other trials.
The weight observation data from the five trials are plotted in Fig. 1, with a separate line for each cage. Rats in the five trials appeared to have similar and expected growth patterns: male rats gained weight faster than the females; the variance among cage weights of the same treatment increased with increasing weight; the rate of weight gain decreased over time. It is also apparent that several cages had below-trend line weights at certain weeks that returned to the trend lines in later weeks. These observations could be considered outliers. We did not remove these outliers, since the original publication of these data sets did not describe any scientific rationale that would justify removing them. Differences in results between otherwise comparable data sets (A vs B or D vs E) may be attributable to these outliers.
Observed weight data for the five feeding studies conducted as part of the European Commission funded GMO Risk Assessment and Communication of Evidence (GRACE) project, where GMO11 and GMO33 represent diets with 11% and 33% GM maize incorporation, Control represents diets including near-isogenic control maize, and Conv, Conv1, and Conv2 represent diets including different varieties of conventional maize
Linear mixed models (LMMs)
Before introducing specific models in this paper, we will review the general theory of LMMs and the nomenclature we will use. A LMM can be expressed in the following matrix equation:
where \({\varvec{y}}\) is the response vector, \({\varvec{\beta}}\) and \({\varvec{u}}\) are the coefficient vectors for fixed effects and random effects, respectively, X and Z are the design matrices for fixed effects and random effects, and \({\varvec{e}}\) is the error vector. We assume \({\varvec{u}}\) and \({\varvec{e}}\) are independent and \({\varvec{u}} \sim N\left( {0,{\varvec{G}}} \right), {\varvec{e}} \sim N\left( {0,{\varvec{R}}} \right)\). Following the common convention, we refer to these as “G-side” and “R-side” random effects and the associated covariance matrices \({\varvec{G}}\) and \({\varvec{R}}\) as “G-side” and “R-side” covariance structures. For additional theoretical discussion of LMMs, interested readers can refer to Vonesh and Chinchilli (1997), Sjoblad et al. (1992), and Littell et al. (2006, pp. 734–756).
Polynomial models with R-side covariance structures
We initially compared polynomial LMMs with linear, quadratic, and cubic responses to time with complex R-side structures. Similar LMMs have often been used to model animal weight growth (Schmidt et al. 2016, 2017; Wang and Goonewardene 2004; Zeljenková et al. 2016). The mathematical formulations of the models we considered are given as follows. Let \(Y_{ijt}\) be the mean body weight of rats in cage \(j\) of treatment \(i\) at week \(t\), for \(i = 1, \ldots ,I, j = 1, \ldots ,J, t = 0, \ldots ,T\), where \(T\) was a value less than 13 for trials A, B, D, and E, to match the actual durations described above. The three models are given as:
where \(\epsilon_{{\varvec{ij}}} = \left( {\epsilon_{ij0} ,\epsilon_{ij1} , \ldots ,\epsilon_{ijT} } \right)^{{\prime }} \sim^{iid} N\left( {0,{\varvec{\Sigma}}} \right)\); thus, the error covariance matrix \({\varvec{R}}\) is a block diagonal with \({\varvec{\Sigma}}\), the repeated measure error covariance matrix at the subject (cage) level, along the diagonal. In the models, \(\alpha\) represents the effect mean at time 0 (fixed intercept) for all treatment groups; \(\beta_{i}\), \(\gamma_{i}\), and \(\delta_{i}\), respectively, represent the slope of the linear, quadratic, and cubic terms (fixed coefficients) of the polynomial response of weight to the time variable for treatment \(i\). Note that though polynomial coefficients were varied among treatment groups, the same intercept value was fit for all treatments, since the intercept defines the initial weight and the diet treatments could not cause effects before the rats were fed. All three models were fitted using Toeplitz structures for \({\varvec{\Sigma}}\). The general form of an \(n \times n\) Toeplitz matrix is provided as follows:
In a Toeplitz covariance structure, all observations have the same variance, and covariance parameters depend on the time lag between observations. Alternative structures for \({\varvec{\Sigma}}\) could also be fit as discussed later.
Polynomial models with G-side random effects
As an alternative to R-side covariance structures such as the Toeplitz structure or heteroscedastic structures, we considered a class of LMMs with G-side random effects, also known as random coefficient models. Random effects represent individual subject (cage) effects as random deviations from the fixed treatment effects, and they allow each cage to have its own trajectory or inherent trend with its own particular features. Random coefficient models were considered for linear, quadratic, and cubic mean response functions labeled as models 2L, 2Q, and 2C. The quadratic function is presented below as an example to illustrate the mathematical set-up of the random coefficient models. We considered the following model, for \(i = 1, \ldots ,I, j = 1, \ldots ,J, t = 0, \ldots T\):
where \(\alpha\), \(\beta_{i}\), \(\gamma_{i}\) denote fixed effects with the same interpretation as described in the prior section titled “Polynomial models with R-side covariance structures” and \(a_{ij}\), \(b_{ij}\), \(g_{ij}\) denote random effects associated with polynomial coefficients under the following assumptions: \(a_{ij} \sim^{iid} N\left( {0,\sigma_{a}^{2} } \right)\), \(b_{ij} \sim^{iid} N\left( {0,\sigma_{b}^{2} } \right)\), \(g_{ij} \sim^{iid} N\left( {0,\sigma_{g}^{2} } \right)\).
All \(a_{ij}\), \(b_{ij}\), and \(g_{ij}\) were modeled as independent from each other, and from \(\epsilon_{ijt}\). Models 2L and 2C were constructed in a similar manner.
Nonlinear mixed models
Nonlinear functions have been used extensively in growth curve modeling (Hammill et al. 1995; Kuhi et al. 2010; Richards 1959) and might be expected to provide better model fit than polynomial models. Here we describe our application of the Gompertz and Richards nonlinear functions evaluated with Toeplitz R-side structures and separately with G-side random coefficients using the same approaches as the polynomial models.
There are many parameterizations of the Gompertz function in the literature, often including a parameter representing the maximum relative growth rate at the inflection point (Lewis et al. 2002; Sugden et al. 1981). In the GRACE studies, the rats were partially grown at the initiation of each study, and as seen in Fig. 1, the rats appear to have already passed their period of fastest growth. When a Gompertz function is fitted to such data, the inflection point is located at a time point prior to the start of the experiment. Thus, estimates of an inflection point parameter for a given treatment group would be difficult to interpret biologically. In order to improve the interpretation of growth curve parameters, we chose the parameterization introduced by Hammill et al. (1995), which is given by:
where \(G\left( {t;\alpha ,\omega ,\kappa } \right)\) is the expected weight at week \(t\), \(\alpha\) is the initial weight at week \(0\), \(\omega\) is the asymptotic final weight, and \(\kappa\) is the growth rate factor, which equals the growth rate at week \(0\), i.e., \(G^{{\prime }} \left( 0 \right) = \kappa\). Note that although \(\kappa\) can be intuitively interpreted as the initial growth rate, \(\kappa\) influences the growth rate at all time points since the expression \(G^{{\prime }} \left( t \right)\) contains \(\kappa\) at all times \(t\).
The Richards function (Richards 1959), also known as the generalized logistic function, is a four-parameter generalization of the Gompertz function. We used the parameterization:
where \(R\left( {t;\alpha ,\omega ,\mu ,\varsigma } \right)\) is the expected weight at week \(t\), \(\alpha\) is the initial weight at week \(0\), \(\omega\) is the asymptotic final weight, \(\mu\) is the growth rate factor (\({\text{week}}^{ - 1}\)), and \(\varsigma\) determines the shape of the curve. Note that \(\mu\) cannot be interpreted as simply as \(\kappa\) in the Gompertz function. For different values of the parameter \(\varsigma\), the Richards function simplifies to the Gompertz (\(\varsigma \to 0)\), logistic (\(\varsigma = 1\)), or monomolecular (\(\varsigma = - 1)\) functions (France and Thornley 1984, p. 155).
Gompertz and Richards function models were fitted in PROC NLMIXED in SAS (Version 9.4). We evaluated two models based on the Gompertz function. Let \(Y_{ijt}\) be the mean weight in cage \(j\) of treatment \(i\) at week \(t\), for \(i = 1, \ldots ,I, j = 1, \ldots ,J, t = 0, \ldots ,T\), then the models are given by:
where \(\alpha\), \(\omega_{i}\), and \(\kappa_{i}\) denote fixed effects with the interpretation given above, \(a_{ij}\), \(o_{ij}\), and \(k_{ij}\) denote random effects, and \(\varvec{\epsilon}_{{\varvec{ij}}} = \left( {\epsilon_{ij0} ,\epsilon_{ij1} , \ldots ,\epsilon_{ijt} } \right)^{\prime } \sim^{iid} N\left( {{\mathbf{0}},{\varvec{\Sigma}}} \right)\), under the following assumptions: \(a_{ij} \sim^{iid} N\left( {0,\sigma_{a}^{2} } \right),\,\,o_{ij} \sim^{iid} N\left( {0,\sigma_{o}^{2} } \right),\,\,k_{ij} \sim^{iid} N\left( {0,\sigma_{k}^{2} } \right)\).
\(a_{ij}\), \(o_{ij}\), and \(k_{ij}\) were modeled as independent from each other and from \(\varepsilon_{ijt}\). Model 1G was fit using the Toeplitz R-side covariance structure for \({\varvec{\Sigma}}\), and a constant variance structure for \({\varvec{\Sigma}}\) was fit for model 2G. Note that \(\alpha\), the initial weight, was assumed to be constant for all treatment groups since the diets could not cause effects before the rats were fed.
We evaluated two models based on Richards function:
where \(\alpha\), \(\omega_{i}\), \(\mu_{i}\), and \(\varsigma\) denote fixed effects with the interpretation given above, \(a_{ij}\), \(o_{ij}\), and \(m_{ij}\) denote random effects, and \(\epsilon_{{\varvec{ij}}} = \left( {\epsilon_{ij0} ,\epsilon_{ij1} , \ldots ,\epsilon_{ijT} } \right)^{\prime} \sim^{iid} N\left( {0,{\varvec{\Sigma}}} \right)\), under the following assumptions: \(a_{ij} \sim^{iid} N\left( {0,\sigma_{a}^{2} } \right),\,\,o_{ij} \sim^{iid} N\left( {0,\sigma_{o}^{2} } \right),\,\,m_{ij} \sim^{iid} N\left( {0,\sigma_{m}^{2} } \right)\).
\(a_{ij}\), \(o_{ij}\), and \(m_{ij}\) were modeled as independent from each other and from \(\epsilon_{ijt}\). Model 1R was fit using the Toeplitz R-side covariance structure for \({\varvec{\Sigma}}\) and a constant variance structure for \({\varvec{\Sigma}}\) was fit for model 2R. Again, \(\alpha\) was assumed to be constant for all treatment groups. In order to simplify the models, we treated \(\varsigma\) as a study-sex-specific parameter with a constant value for all cages in all treatment groups.
Model comparisons
The models were evaluated for each study by visual inspection of simulation and residual plots and by comparison of information criteria. The MIXED and NLMIXED procedures in SAS (Version 9.4) were used to fit models to the data and generate studentized residual plots. Once a model was fit, we assumed the fitted model with the estimated parameters was the data generating mechanism for the raw data and simulated a data set based on this mechanism. Observations at each time point were simulated and plotted for a number of cages for each treatment equal to the quantities tested in the study. Features of the simulated dataset that substantially differed from the raw data could indicate flaws in the model’s approximation of the true data generating mechanism. Note that each simulation plot is just one possible realization of the fitted model, and may include random effects or error values from the extremes of the distributions simulated. The same can be said of the raw data. Thus, limited emphasis should be placed on interpretation of a single curve or observation that does not match between the raw and simulated data. Despite these limitations, the plotted simulation is a useful diagnostic tool that allows for intuitive assessment and comparisons among models. We used the maximum likelihood method to estimate model parameters, since use of the restricted maximum likelihood (REML) method would have made the information criteria non-comparable among the methods (Littell et al. 2006, pp. 752-754).
Comparisons among the models were made after being fit separately to the ten data sets consisting of male and female data from the five trials. Results from all five trials can be found in the supplemental material. Since results were generally similar among the ten data sets, the results from the female trial A data set are presented and discussed, as this trial was of intermediate size among the five and the data do not include any obvious outliers. Any differences in results among the sexes or trials are discussed.
Results and discussion
Polynomial models with R-side covariance structures
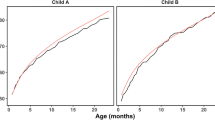
A comparison between the graphs of raw data and simulated data for model 1L suggests that a strictly linear relation failed to capture the nonlinear relationship between weight and week variables (Fig. 2). The data variance not captured by the model resulted in larger estimates of error variance, resulting in wide separation among the lines in the simulation based on model 1L. The graphs of models 1Q and 1C show an improvement over the linear model in their approximation of the observed data. In all three models, the Toeplitz structure assumed homogeneous variance across weeks and thus did not capture the heteroscedasticity apparent in the raw data. The simulated data graphs for the other data sets suggest the same conclusions (Supplemental Fig. 4, 7, 10, 13, 16, 19, 22, 25, and 28).
Female trial A raw data (leftmost) and data simulated assuming the generating mechanism matched the respective model and estimated parameters, where GMO11 and GMO33 represent diets with 11% and 33% GM maize incorporation, Control represents diets including near-isogenic control maize, and Conv, Conv1, and Conv2 represent diets including different varieties of conventional maize
In addition to the simulation plots, plots of studentized residuals against predicted values were produced for models 1L, 1Q and 1C; the residuals for the female data from trial A are shown in Fig. 3. The residual plots helped to determine if the assumptions necessary for each model were met. Ideally, the residual values should be evenly spread vertically across the horizontal zero line with no pattern, indicating independence, and the amount of spread should be constant for all predicted values, indicating homoscedasticity. These residual plots reflect similar model behavior as the simulation plots. The curvature of the pattern of residuals from the linear model indicates that it did not adequately model the nonlinear relationship. The quadratic and cubic models showed more symmetric patterns about the zero line. The triangular shapes in the residual plots for models 1Q and 1C indicate that the Toeplitz structure failed to capture the heteroscedasticity across weeks. This heteroscedasticity was also present for most of the other datasets (Supplemental Fig. 6, 9, 12, 15, 18, 21, 24, 27, and 30).
Information criteria were also used to compare the three models, as shown in Table 1. The relative differences in values among the models agreed with the simulation and residual plots. That is, the quadratic model substantially improved model fit, and the cubic resulted in a slight improvement over the quadratic. Information criteria from the other data sets resulted in the same rank ordering of the models, but the magnitude of difference varied (Supplemental Tables 1 to 10).
The failure of these models to capture heteroscedasticity in the data might hamper their use to accurately estimate the relationship between time and weight for a particular treatment or to identify differences among diets (Littell et al. 2006, p. 794; Scheffé 1959). Simulations by Ferron et al. (2002) demonstrated that misspecification of the error covariance structure in growth curves led to biased estimates of variance parameters. One may consider fitting a heteroscedastic covariance structure, by allowing variance heterogeneity in the Toeplitz structure or another structure. Cubic models with compound symmetric and AR(1) heterogeneous variance structures were applied to all ten data sets and residuals were plotted against predicted values (Supplemental Fig. 31 to 40). Unresolvable convergence issues occurred for most of the data sets when a heteroscedastic Toeplitz structure was attempted. In our evaluations of these heterogeneous variance structures, we observed moderate to extreme biases in the fixed effect estimates appearing as uneven spread of residuals about the zero line.
Polynomial models with G-side random effects
Simulation plots, residual plots, and information criteria were compared for models 2L (linear), 2Q (quadratic), and 2C (cubic). Results for the female trial A data set are shown in Figs. 4, 5, and Table 2.
As shown in Fig. 4, the random coefficient models resulted in increasing separation among simulated growth curves of each cage over time. This better reflects the raw data pattern as compared to the models with Toeplitz error covariance structures. As observed with the models including R-side structures, model 2L was overly simplistic, whereas the quadratic and cubic models better captured the curvature observed in the raw data. Results for the other data sets were similar (Supplemental Fig. 5, 8, 11, 14, 17, 20, 23, 26, and 29).
The studentized residuals from the linear and quadratic models applied to female trial A data showed asymmetric patterns (Fig. 5). These patterns indicate that these models did not adequately fit the data, whereas the cubic model performed better. When applied to the other data sets, the cubic model resulted in modest to substantial improvements in model fit (Supplemental Fig. 6, 9, 12, 15, 18, 21, 24, 27, and 30). However, the residuals based on the cubic model still showed asymmetry for the female data from trial C and the male data from trials A, C, D, and E.
The information criteria (Table 2) indicate that the higher order polynomials led to better performance for the female trial A data. Comparing Tables 1 and 2, the information criteria for the three random coefficient models were uniformly greater (indicating worse fit) than those of the corresponding Toeplitz structure models with smaller differences for higher order polynomials. The same pattern of results was observed for both sexes in trials A, B, and C, and male data in trial D (Supplemental Tables 1 to 6 and 8). For the remaining three data sets from trials D and E, the information criteria for random coefficient cubic models were slightly better than those for model 1C. Trials D and E included fewer treatments and cages per diet, suggesting that the Toeplitz R-side structure may require more data to provide better model fit than G-side random coefficients when used with fairly complex (cubic) polynomial models.
The model comparisons above demonstrated that while easily interpreted, LMMs with only a single slope parameter per treatment were insufficient to capture the growth curve pattern in the raw data, regardless of whether an R-side or G-side covariance structure was used. When random coefficients were used in the model, substantial improvements were observed with the higher order polynomials. However, the asymmetric spread of residuals for model 2C suggested that the mean response function could be improved beyond the cubic. This was not apparent for model 1C, because the R-side covariance structure captured the deviations from the cubic curve as correlated random error.
Neither random coefficient models nor Toeplitz R-side structure models were indisputably superior with any of the polynomial parameterizations evaluated. Likely due to the limited number of observations in these experiments, convergence could not be achieved for models attempted with both Toeplitz error covariance structures and random coefficients. Models combining random coefficients and simpler error covariance structures are a potential option for further optimization which we did not pursue. Besides the graphs and information criteria, some additional considerations favor the random coefficient models. First, the random coefficient models required fewer parameters than the LMM with Toeplitz structures. This difference would increase with more time points, since the Toeplitz structure requires the same number of parameters as time points modeled. Additionally, random coefficients allow each cage to be modeled with its own growth curve. Such models may better reflect biological reality than using the Toeplitz structure which models curvature differences among experimental units as correlated errors.
Nonlinear mixed models
As described for the polynomial models, the nonlinear models were compared using simulation plots (Fig. 6), residual plots (Fig. 7), and information criteria (Table 3). For data of both sexes from trials C, D, and E and male data from trial B, models 1G and 1R encountered numerical or convergence issues that could not be resolved. Viewing the simulation plots for the female trial A data, it can be observed that models 1G and 1R failed to capture the heteroscedasticity over time, and the same results occurred with the other data sets for which these models converged (Supplemental Fig. 4 and 7).
As with the simulation plots, the residual plots show evidence that the Toeplitz models failed to account for heteroscedasticity. The residuals of model 2G form an obvious curved pattern in the plot, indicating that the Gompertz function failed to capture the full complexity of the growth curve (Fig. 7). The residuals show little to no curvature in models 2R, 1G, and 1R, with decreasing curvature in that order, indicating that these models are capturing most or all of the relationships between time and weight. Both random effects models fit to male data from trials D and E resulted in curvatures in residual plots due to relatively small growth rates in the beginning weeks for these two data sets (Supplemental Fig. 24 and 30). Furthermore, for the other eight data sets, models 2R, 1G, and 1R displayed little to no curvature for each data set successfully fit, whereas model 2G had curvature for all but the female data of trials D and E.
The Richards function resulted in similar or better information criteria values as compared to the other functions evaluated. For the successfully fit Toeplitz models, the Gompertz and Richards functions resulted in similar or slightly better model fit scores compared to the cubic function. When random coefficients were used, the Richards function resulted in substantially lower information criteria values than the Gompertz and cubic models. The Toeplitz models resulted in better model fit based on information criteria values as compared to the corresponding random coefficient models for most data sets. The exception was Richards function which resulted in lower information criteria with random coefficients for two of the three data sets where the Toeplitz version was successfully fit. It is important to recall that Toeplitz models failed to account for heteroscedasticity as demonstrated in the residual diagnostic plots. This suggests that when the underlying mean response function poorly captures the overall growth pattern, the correlations in the Toeplitz error structure can capture the deviations from the mean growth pattern at each time point. However, when an adequate function (Richards) is chosen to model the mean growth pattern, random coefficients can efficiently model variability among individual cages and effectively account for heteroscedasticity over time.
From a practical standpoint, we note that the Toeplitz structure was difficult to implement with the nonlinear functions. For the majority of the data sets, no starting values could be found to allow model convergence and properly conditioned final Hessian matrices for models 1G and 1R, even with repeated attempts. Additionally, it took much longer to fit the Toeplitz models compared to the corresponding random coefficients models using the NLMIXED procedure. The complexity of the Toeplitz structure could be even more problematic for data sets with more time points, since the number of parameters in the Toeplitz structure equals the number of time points.
The preceding discussion evaluated various aspects of modeling 90-day rat studies including mean response function (polynomials and nonlinear growth curves), cage to cage variability structure (G-side random effects), and within cage error (R-side random effects). The options evaluated are not exhaustive. Some additional options include G-side structures that do not assume independence of the random factors and simpler R-side structures combined with G-side structures. Although Model 2R appeared to be the best model choice for most of the data sets we evaluated, it would not be appropriate to conclude that it should always be chosen for the analysis of 90-day rat studies. Instead, it would be advisable for researchers to attempt multiple models of greater and lesser complexity, choosing the one with superior information criteria scores and residual plots that do not show major flaws.
Treatment comparisons
When modeling body weight data from 90-day rodent feeding studies, the goal is to estimate and compare the effects of diet treatments on rodent growth. Once a model is chosen and fit to the data, the parameters for that model can be estimated specifically for each diet. Confidence intervals for the parameter estimates can be provided to establish the accuracy of those estimates. Approximate F-tests or t-tests can be used to evaluate the statistical significance of hypotheses of interest. Conducting hypothesis tests on parameters that apply to the whole growth curve reduces the risk of multiplicity as compared to separately comparing treatment effects at each time point.
The datasets from the GRACE project would not be expected to show any true effects of the GM diets on rat growth, since extensive evaluations beyond weight measurements were previously conducted and showed no biologically relevant effects of the GM diets (Schmidt et al. 2017; Zeljenková et al. 2016, 2014). Therefore, any observed treatment effects for these datasets would be false positives. In analyses of other datasets, interpretation of statistically significant treatment comparisons would require expert evaluation within the context of all the results of the study, as statistical significance does not necessarily or automatically imply biological relevance (EFSA 2011c).
The scope of this investigation prevents us from evaluating the ability of these models to detect true adverse effects in terms of growth rates or final 13th week weights. Such an evaluation would require analysis of feeding trials with positive controls that have known biologically relevant effects on growth rate and/or final weight. Once biologically relevant effect sizes are determined, simulations could be conducted to compare these models in terms of statistical power and false-positive error rates. Since deviations from assumptions generally lead to loss of statistical power or increased false-positive rates, it is likely that similar models would be preferred.
Conclusions
For body weight data from the five 90-day rat feeding trials conducted as part of the GRACE project, models using the Richards function and random coefficients were generally superior to the other models evaluated and dramatically improved upon one-parameter linear models with complex error structures. In terms of model fit, the complex Toeplitz error variance structure compensated for overly simple models of the relationship between time and weight. However, these models may be incorrectly modeling true complexity in the relationship between time and weight as error correlation. Additionally, the Toeplitz models did not effectively model the observed heteroscedasticity over time. Random coefficient models captured heteroscedasticity over time while partitioning out variation among individual cages with modest increases in parameter numbers. Inferior model fit was observed when random coefficients were used with simple functions. Only when a sufficiently complex function was used could superior model fit be achieved along with the benefits of random coefficients. The Richards function with random coefficients was well suited for most of the data sets analyzed, but may not be optimal for other growth curve data sets. Other investigators should take care to observe the residual plots from applying this model to ensure that it adequately captures the growth pattern. Application of these models to make comparisons among diet treatments will also require expert evaluation of biological relevance within the context of all study results.
This paper focused on evaluations of methods for statistical analysis of body weight data from studies that follow the EFSA guidance. However, similar model evaluations could be made for other animal feeding studies, and some of the same approaches or conclusions may apply.
References
EFSA (2011a) Guidance for risk assessment of food and feed from genetically modified plants. EFSA J 9:2150. https://doi.org/10.2903/j.efsa.2011.2150
EFSA (2011b) Guidance on conducting repeated-dose 90-day oral toxicity study in rodents on whole food/feed. EFSA J 9:2438. https://doi.org/10.2903/j.efsa.2011.2438
EFSA (2011c) Statistical Significance and biological relevance. EFSA J 9:2372. https://doi.org/10.2903/j.efsa.2011.2372
Ferron J, Dailey R, Yi Q (2002) Effects of misspecifying the first-level error structure in two-level models of change. Multivar Behav Res 37:379–403. https://doi.org/10.1207/S15327906MBR3703_4
France J, Thornley JHM (1984) Mathematical models in agriculture: a quantitative approach to problems in agriculture and related sciences. Butterworths, London
Hammill M, Kingsley M, Beck G, Smith T (1995) Growth and condition in the Northwest Atlantic harp seal. Can J Fish Aquat Sci 52:478–488. https://doi.org/10.1139/f95-049
ILSI Task Force (2004) Nutritional and safety assessments of foods and feeds nutritionally improved through biotechnology: an executive summary a task force report by the International Life Sciences Institute, Washington, D.C. Compr Rev Food Sci Food Saf 3:35–104. https://doi.org/10.1111/j.1541-4337.2004.tb00059.x
Kuhi HD, Porter T, Lopez S, Kebreab E, Strathe A, Dumas A, Dijkstra J, France J (2010) A review of mathematical functions for the analysis of growth in poultry. World’s Poult Sci J 66:227–240. https://doi.org/10.1017/S0043933910000280
Lewis RW, Billington R, Debryune E, Gamer A, Lang B, Carpanini F (2002) Recognition of adverse and nonadverse effects in toxicity studies. Toxicol Pathol 30:66–74. https://doi.org/10.1080/01926230252824725
Littell RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O (2006) Example: variance component estimates equal to zero. In: SAS® for mixed models, 2 edn. SAS Institute Inc., Cary
OECD (2003) Considerations for the safety assessment of animal feedstuffs derived from genetically modified plants. Organisation for Economic Co-operation and Development, ENV/JM/MONO(2003)10, Paris
Richards F (1959) A flexible growth function for empirical use. J Exp Bot 10:290–301. https://doi.org/10.1093/jxb/10.2.290
Scheffé H (1959) The analysis of variance. Wliey, New York
Schmidt K, Schmidtke J, Kohl C, Wilhelm R, Schiemann J, van der Voet H, Steinberg P (2016) Enhancing the interpretation of statistical P values in toxicology studies: implementation of linear mixed models (LMMs) and standardized effect sizes (SESs). Arch Toxicol 90:731–751. https://doi.org/10.1007/s00204-015-1487-8
Schmidt K, Schmidtke J, Schmidt P, Kohl C, Wilhelm R, Schiemann J, van der Voet H, Steinberg P (2017) Variability of control data and relevance of observed group differences in five oral toxicity studies with genetically modified maize MON810 in rats. Arch Toxicol 91:1977–2006. https://doi.org/10.1007/s00204-016-1857-x
Sjoblad RD, McClintock JT, Engler R (1992) Toxicological considerations for protein components of biological pesticide products. Regul Toxicol Pharmacol 15:3–9. https://doi.org/10.1016/0273-2300(92)90078-N
Sugden LG, Driver E, Kingsley MC (1981) Growth and energy consumption by captive mallards. Can J Zool 59:1567–1570. https://doi.org/10.1139/z81-213
Vonesh E, Chinchilli VM (1997) Linear and nonlinear models for the analysis of repeated measurements. Marcel Dekker, New York
Wang Z, Goonewardene LA (2004) The use of MIXED models in the analysis of animal experiments with repeated measures data. Can J Anim Sci 84:1–11. https://doi.org/10.4141/A03-123
Zeljenková D, Ambrušová K, Bartušová M, Kebis A, Kovrižnych J, Krivošíková Z, Kuricová M, Líšková A, Rollerová E, Spustová V, Szabová E, Tulinská J, Wimmerová S, Levkut M, Révajová V, Sevčíková Z, Schmidt K, Schmidtke J, La Paz JL, Corujo M, Pla M, Kleter GA, Kok EJ, Sharbati J, Hanisch C, Einspanier R, Adel-Patient K, Wal J-M, Spök A, Pöting A, Kohl C, Wilhelm R, Schiemann J, Steinberg P (2014) Ninety-day oral toxicity studies on two genetically modified maize MON810 varieties in Wistar Han RCC rats (EU 7th Framework Programme project GRACE). Arch Toxicol 88:2289–2314. https://doi.org/10.1007/s00204-014-1374-8
Zeljenková D, Aláčová R, Ondrejková J, Ambrušová K, Bartušová M, Kebis A, Kovrižnych J, Rollerová E, Szabová E, Wimmerová S (2016) One-year oral toxicity study on a genetically modified maize MON810 variety in Wistar Han RCC rats (EU 7th Framework Programme project GRACE). Arch Toxicol 90:2531–2562. https://doi.org/10.1007/s00204-016-1798-4
Acknowledgements
We would like to acknowledge Bonnie Hong for initiating this project and George Milliken for his important early work on this project. We also thank them for providing valuable reviews and suggestions.
Author information
Authors and Affiliations
Contributions
KH was responsible for statistical analysis. CW was responsible for manuscript preparation. Both authors contributed ideas and research.
Corresponding author
Ethics declarations
Conflict of interest
During the conduct of the study, KH was employed by Iowa State University and contracted by DuPont Pioneer, and CW was employed by DuPont Pioneer. The work was funded by DuPont Pioneer (now Corteva Agriscience).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Huang, K., Walker, C.A. Comparisons of statistical models for growth curves from 90-day rat feeding studies. Arch Toxicol 93, 2397–2408 (2019). https://doi.org/10.1007/s00204-019-02496-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-019-02496-5