Abstract
Key message
Using a much higher number of SNP markers and larger sample sizes than all the previous studies, we characterized the genetic relationships among wild and cultivated plants of section Beta.
Abstract
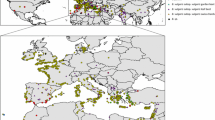
We analyzed the genetic variation of Beta section Beta, which includes wild taxa (Beta macrocarpa, B. patula, B. vulgaris subsp. adanensis and B. vulgaris subsp. maritima) and cultivars (fodder beet, sugar beet, garden beet, leaf beet, and swiss chards), using 9724 single nucleotide polymorphism markers. The analyses conducted at the individual level without a priori groups confirmed the strong differentiation of B. macrocarpa and B. vulgaris subsp. adanensis from the other taxa. B. vulgaris subsp. maritima showed a complex genetic structure partly following a geographical pattern, which confounded the differences between this taxon and the cultivated varieties. Cultivated varieties were structured into three main groups: garden beets, fodder and sugar beets, and leaf beets and swiss chards. The genetic structure described here will be helpful to correctly estimate linkage disequilibrium and to test for statistical associations between genetic markers and environmental variables.


Similar content being viewed by others
References
Abe J, Tsuda C (1987) Genetic analysis for isozyme variation in the section Vulgares, genus Beta. Jpn J Breed 37:253–261
Abe J, Yoshikawa H, Tsuda C (1986) Reproductive barriers in sugar beet (Beta vulgaris) and its wild relatives of the section Vulgares, the genus Beta. J Fac Agric Hokkaido Univ Jpn 63:40–48
Adetunji I, Willems G, Tschoep H et al (2014) Genetic diversity and linkage disequilibrium analysis in elite sugar beet breeding lines and wild beet accessions. Theor Appl Genet 127:559–571. doi:10.1007/s00122-013-2239-x
Andrello M, Henry K, Devaux P et al (2016) Taxonomic, spatial and adaptive genetic variation of Beta section Beta. Theor Appl Genet 129:257–271. doi:10.1007/s00122-015-2625-7
Arnaud J-F, Viard F, Delescluse M, Cuguen J (2003) Evidence for gene flow via seed dispersal from crop to wild relatives in Beta vulgaris (Chenopodiaceae): consequences for the release of genetically modified crop species with weedy lineages. Proc R Soc Lond B Biol Sci 270:1565–1571
Arnaud J-F, Fénart S, Godé C et al (2009) Fine-scale geographical structure of genetic diversity in inland wild beet populations. Mol Ecol 18:3201–3215. doi:10.1111/j.1365-294X.2009.04279.x
Bauchet G, Munos S, Sauvage C et al (2014) Genes involved in floral meristem in tomato exhibit drastically reduced genetic diversity and signature of selection. BMC Plant Biol 14:1
Biancardi E, Panella LW, Lewellen RT (2012) Beta maritima. The origin of beets. Springer, New York
Bonavent J-F, Bessone L, Geny A et al (1989) A possible origin for the sugar beet cytoplasmic male sterility source Owen. Genome 32:322–327. doi:10.1139/g89-448
Buttler KP (1977) Variation in wild populations of annual beet (Beta, Chenopodiaceae). Plant Syst Evol 128:123–136
Campbell LG (2010) Registration of seven sugarbeet germplasms selected from crosses between cultivated sugarbeet and wild species. J Plant Regist 4:149. doi:10.3198/jpr2009.11.0673crg
Cheng D, Kitazaki K, Xu D et al (2009) The distribution of normal and male-sterile cytoplasms in Chinese sugar-beet germplasm. Euphytica 165:345–351. doi:10.1007/s10681-008-9796-0
Cheng D, Yoshida Y, Kitazaki K et al (2011) Mitochondrial genome diversity in Beta vulgaris L. ssp. vulgaris (leaf and garden beet groups) and its implications concerning the dissemination of the crop. Genet Resour Crop Evol 58:553–560. doi:10.1007/s10722-010-9598-9
Curk F, Ancillo G, Ollitrault F et al (2015) Nuclear species-diagnostic SNP markers mined from 454 amplicon sequencing reveal admixture genomic structure of modern citrus varieties. PLoS One 10:e0125628
Desplanque B, Boudry P, Broomberg K et al (1999) Genetic diversity and gene flow between wild, cultivated and weedy forms of Beta vulgaris L. (Chenopodiaceae), assessed by RFLP and microsatellite markers. Theor Appl Genet 98:1194–1201
Dohm JC, Minoche AE, Holtgräwe D et al (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505:546–549. doi:10.1038/nature12817
Draycott AP (ed) (2006) Sugar beet. Blackwell, Oxford
Ecke W, Michaelis G (1990) Comparison of chloroplast and mitochondrial DNA from five morphologically distinct Beta vulgaris cultivars: sugar beet, fodder beet, beet root, foliage beet, and Swiss chard. TAG Theor Appl Genet Theor Angew Genet 79:440–442. doi:10.1007/BF00226149
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor Appl Genet 92:832–839. doi:10.1007/BF00221895
Excoffier L, Dupanloup I, Huerta-Sánchez E et al (2013) Robust demographic inference from genomic and SNP data. PLoS Genet 9:e1003905. doi:10.1371/journal.pgen.1003905
Fénart S, Arnaud J-F, De Cauwer I, Cuguen J (2008) Nuclear and cytoplasmic genetic diversity in weed beet and sugar beet accessions compared to wild relatives: new insights into the genetic relationships within the Beta vulgaris complex species. Theor Appl Genet 116:1063–1077. doi:10.1007/s00122-008-0735-1
Fievet V, Touzet P, Arnaud J-F, Cuguen J (2007) Spatial analysis of nuclear and cytoplasmic DNA diversity in wild sea beet (Beta vulgaris ssp. maritima) populations: do marine currents shape the genetic structure? Mol Ecol 16:1847–1864. doi:10.1111/j.1365-294X.2006.03208.x
Fischer HE (1989) Origin of the “Weisse Schlesische Rübe” (white Silesian beet) and resynthesis of sugar beet. Euphytica 41:75–80. doi:10.1007/BF00022414
Frese L (2010) Conservation and Access to Sugarbeet Germplasm. Sugar Tech 12:207–219. doi:10.1007/s12355-010-0054-0
Frese L, Nachtigall M, Enders M, Pinheiro De Carvalho MAA (2012) Beta patula (Aiton): Genetic diversity analysis. In: Maxted N (ed) \Agrobiodiversity conservation: securing the diversity of crop wild relatives and landraces. CABI, Wallingford
Fritzsche K, Metzlaff M, Melzer R, Hagemann R (1987) Comparative restriction endonuclease analysis and molecular cloning of plastid DNAs from wild species and cultivated varieties of the genus Beta (L.). Theor Appl Genet 74:589–594. doi:10.1007/BF00288857
Hammer K, Stanarius A, Kühne T (1990) Differential occurrence of beet cryptic viruses—a new tool for germplasm characterization and evolutionary studies in beets? Euphytica 45:23–27. doi:10.1007/BF00032145
Hjerdin A, Sall T, Nilsson NO et al (1994) Genetic variation among wild and cultivated beets of the section Beta as revealed by RFLP analysis. J Sugar Beet Res 31:59–67
Hoban S, Kelley JL, Lotterhos KE et al (2016) Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am Nat 188:379–397. doi:10.1086/688018
Hyten DL, Song Q, Zhu Y et al (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci 103:16666–16671. doi:10.1073/pnas.0604379103
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. doi:10.1093/bioinformatics/btr521
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jones PD, Lister DH, Jaggard KW, Pidgeon JD (2003) Future climate impact on the productivity of sugar beet (Beta vulgaris L.) in Europe. Clim Change 58:93–108
Jung C, Pillen K, Frese L et al (1993) Phylogenetic relationships between cultivated and wild species of the genus Beta revealed by DNA “fingerprinting”. Theor Appl Genet 86:449–457. doi:10.1007/BF00838560
Kadereit G, Hohmann S, Kadereit JW (2006) A synopsis of Chenopodiaceae subfam. Betoideae and notes on the taxonomy of Beta. Willdenowia 36:9–19. doi:10.3372/wi.36.36101
Kilian B, Graner A (2012) NGS technologies for analyzing germplasm diversity in genebanks. Brief Funct Genom 11:38–50. doi:10.1093/bfgp/elr046
Kishima Y, Mikami T, Hirai A et al (1987) Beta chloroplast genomes: analysis of fraction I protein and chloroplast DNA variation. Theor Appl Genet. doi:10.1007/BF00262497
Letschert JPW (1993) Beta section Beta: biogeographical patterns of variation, and taxonomy. Wagening Agric Univ Pap 93(1):1–155
Leys M, Petit EJ, El-Bahloul Y et al (2014) Spatial genetic structure in Beta vulgaris subsp. maritima and Beta macrocarpa reveals the effect of contrasting mating system, influence of marine currents, and footprints of postglacial recolonization routes. Ecol Evol 4:1828–1852. doi:10.1002/ece3.1061
Litwiniec A, Gośka M, Choińska B et al (2016) Evaluation of rhizomania-resistance segregating sequences and overall genetic diversity pattern among selected accessions of Beta and Patellifolia. Potential implications of breeding for genetic bottlenecks in terms of rhizomania resistance. Euphytica 207:685–706. doi:10.1007/s10681-015-1570-5
Lu Y, Yan J, Guimarães CT et al (2009) Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms. Theor Appl Genet 120:93–115. doi:10.1007/s00122-009-1162-7
Mangin B, Siberchicot A, Nicolas S et al (2012) Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity 108:285–291. doi:10.1038/hdy.2011.73
Mangin B, Sandron F, Henry K et al (2015) Breeding patterns and cultivated beets origins by genetic diversity and linkage disequilibrium analyses. Theor Appl Genet 128:2255–2271. doi:10.1007/s00122-015-2582-1
Mason AS, Zhang J, Tollenaere R et al (2015) High-throughput genotyping for species identification and diversity assessment in germplasm collections. Mol Ecol Resour 15:1091–1101. doi:10.1111/1755-0998.12379
Maxted N, Ford-Lloyd BV, Jury S et al (2006) Towards a definition of a crop wild relative. Biodivers Conserv 15:2673–2685. doi:10.1007/s10531-005-5409-6
McGrath JM, Derrico CA, Yu Y (1999) Genetic diversity in selected, historical US sugarbeet germplasm and Beta vulgaris ssp. maritima. Theor Appl Genet 98:968–976
Mikami T, Kishima Y, Sugiura M, Kinoshita T (1984) Chloroplast DNA diversity in the cytoplasms of sugar beet and its related species. Plant Sci Lett 36:231–235. doi:10.1016/0304-4211(84)90174-3
Mikami T, Yamamoto MP, Matsuhira H et al (2011) Molecular basis of cytoplasmic male sterility in beets: an overview. Plant Genet Resour 9:284–287. doi:10.1017/S1479262111000177
Mita G, Dani M, Casciari P et al (1991) Assessment of the degree of genetic variation in beet based on RFLP analysis and the taxonomy of Beta. Euphytica 55:1–6. doi:10.1007/BF00022552
Monteiro F, Romeiras MM, Batista D, Duarte MC (2013) Biodiversity assessment of sugar beet species and its wild relatives: linking ecological data with new genetic approaches. Am J Plant Sci 04:21–34. doi:10.4236/ajps.2013.48A003
Morin PA, Luikart G, Wayne RK, The SNP workshop group (2004) SNPs in ecology, evolution and conservation. Trends Ecol Evol 19:208–216. doi:10.1016/j.tree.2004.01.009
Nagamine T, Catty JP, Ford-Lloyd BV (1989) Phenotypic polymorphism and allele differentiation of isozymes in fodder beet, multigerm sugar beet and monogerm sugar beet. Theor Appl Genet 77:711–720
Ober ES, Clark CJA, Bloa ML et al (2004) Assessing the genetic resources to improve drought tolerance in sugar beet: agronomic traits of diverse genotypes under droughted and irrigated conditions. Field Crops Res 90:213–234. doi:10.1016/j.fcr.2004.03.004
Rong J, Lammers Y, Strasburg JL et al (2014) New insights into domestication of carrot from root transcriptome analyses. BMC Genom 15:1
Saccomani M, Stevanato P, Trebbi D et al (2009) Molecular and morpho-physiological characterization of sea, ruderal and cultivated beets. Euphytica 169:19–29. doi:10.1007/s10681-009-9888-5
Santoni S, Bervillé A (1992a) Characterization of the nuclear ribosomal DNA units and phylogeny of Beta L. wild forms and cultivated beets. Theor Appl Genet 83:533–542
Santoni S, Bervillé A (1992b) Two different satellite DNAs in Beta vulgaris L.: evolution, quantification and distribution in the genus. Theor Appl Genet 84:1009–1016
Santoni S, Bervillé A (1992c) Evidence for gene exchanges between sugar beet (Beta vulgaris L.) and wild beets: consequences for transgenic sugar beets. Plant Mol Biol 20:578–580
Santoni S, Faivre-Rampant P, Moreau E, Bervillé A (1991) Rapid control of purity for the cytoplasm of male-sterile seed stocks by means of a dot hybridization assay. Mol Cell Probes 5:1–9
Schiffels S, Durbin R (2014) Inferring human population size and separation history from multiple genome sequences. Nat Genet 46:919–925. doi:10.1038/ng.3015
Schoville SD, Bonin A, François O et al (2012) Adaptive genetic variation on the landscape: methods and cases. Annu Rev Ecol Evol Syst 43:23–43. doi:10.1146/annurev-ecolsys-110411-160248
Senda M, Onodera Y, Mikami T (1998) Cytoplasmic diversity in leaf beet cultivars as revealed by mitochondrial DNA analysis. Hereditas 128:127–132
Shen Y, Ford-Lloyd BV, Newbury J (1998) Genetic relationships within the genus Beta determined using both PCR-based marker and DNA sequencing techniques. Heredity 80:624–632
Shun ZF, Chu SY, Frese L (2000) Study on the relationship between Chinese and East Mediterranean Beta vulgaris L. subsp. vulgaris (leaf beet group) accessions. In: Maggioni L, Frese L, Germeier C, Lipman E (eds) Report of a working group on Beta. International Plant Genetic Resources Institute, Rome, pp 65–69
Stevanato P, Trebbi D, Biancardi E et al (2013) Evaluation of genetic diversity and root traits of sea beet accessions of the Adriatic Sea coast. Euphytica 189:135–146. doi:10.1007/s10681-012-0775-0
Thermo Fisher Scientific Inc. (2017) AxiomTM Genotyping Solution Data Analysis Guide. http://tools.thermofisher.com/content/sfs/manuals/axiom_genotyping_solution_analysis_guide.pdf. Accessed 23 March 2017
Tian H-L, Wang F-G, Zhao J-R et al (2015) Development of maizeSNP3072, a high-throughput compatible SNP array, for DNA fingerprinting identification of Chinese maize varieties. Mol Breed. doi:10.1007/s11032-015-0335-0
van Geyt JPC, Lange W, Oleo M, De Bock TSM (1990) Natural variation within the genus Beta and its possible use for breeding sugar beet : a review. Euphytica 49:57–76
Viard F, Arnaud J-F, Delescluse M, Cuguen J (2004) Tracing back seed and pollen flow within the crop-wild Beta vulgaris complex: genetic distinctiveness vs. hot spots of hybridization over a regional scale. Mol Ecol 13:1357–1364. doi:10.1111/j.1365-294X.2004.02150.x
Villain S (2007) Histoire évolutive de la section Beta: mise en évidence des phénomènes d’hybridation et de spéciation au sein de la Section dans le bassin Méditerranéen. Ph.D., Université Lille 1-Sciences et Technologies
Wang M, Goldman IL (1999) Genetic distance and diversity in table beet and sugar beet accessions measured by randomly amplified polymorphic DNA. J Am Soc Hortic Sci 124:630–635
Warschefsky E, Penmetsa RV, Cook DR, von Wettberg EJB (2014) Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am J Bot 101:1791–1800. doi:10.3732/ajb.1400116
Weihe A, Dudareva NA, Veprev SG et al (1991) Molecular characterization of mitochondrial DNA of different subtypes of male-sterile cytoplasms of the sugar beet Beta vulgaris L. Theor Appl Genet 82:11–16. doi:10.1007/BF00231271
Winkler LR, Michael Bonman J, Chao S et al (2016) Population structure and genotype–phenotype associations in a collection of oat landraces and historic cultivars. Front Plant Sci. doi:10.3389/fpls.2016.01077
Würschum T (2012) Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet 125:201–210. doi:10.1007/s00122-012-1887-6
Würschum T, Maurer HP, Kraft T et al (2011) Genome-wide association mapping of agronomic traits in sugar beet. Theor Appl Genet 123:1121–1131. doi:10.1007/s00122-011-1653-1
Yoshida Y, Matsunaga M, Cheng D et al (2012) Mitochondrial minisatellite polymorphisms in fodder and sugar beets reveal genetic bottlenecks associated with domestication. Biol Plant 56:369–372. doi:10.1007/s10535-012-0101-7
Acknowledgements
This study was funded by the French Government, under the management of the Research National Agency (ANR-11-BTBR-0007) through the AKER program (included in the “Investissements d’avenir”).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Compliance with ethical standards
Not applicable.
Additional information
Communicated by André J. Bervillé.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Andrello, M., Henry, K., Devaux, P. et al. Insights into the genetic relationships among plants of Beta section Beta using SNP markers. Theor Appl Genet 130, 1857–1866 (2017). https://doi.org/10.1007/s00122-017-2929-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2929-x