Abstract
Key message
The RpsQ Phytophthora resistance locus was finely mapped to a 118-kb region on soybean chromosome 3. A best candidate gene was predicted and three co-segregating gene markers were developed.
Abstract
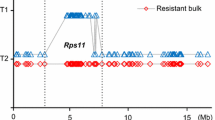
Phytophthora root rot (PRR), caused by Phytophthora sojae, is a major threat to sustainable soybean production. The use of genetically resistant cultivars is considered the most effective way to control this disease. The Chinese soybean cultivar Qichadou 1 exhibited a broad spectrum resistance, with a distinct resistance phenotype, following inoculation with 36 Chinese P. sojae isolates. Genetic analyses indicated that the disease resistance in Qichadou 1 is controlled by a single dominant gene. This gene locus was designated as RpsQ and mapped to a 118-kb region between BARCSOYSSR_03_0165 and InDel281 on soybean chromosome 3, and co-segregated with Insert11, Insert144 and SNP276. Within this region, there was only one gene Glyma.03g27200 encoding a protein with a typical serine/threonine protein kinase structure, and the expression pattern analysis showed that this gene induced by P. sojae infection, which was suggested as a best candidate gene of RpsQ. Candidate gene specific marker Insert144 was used to distinguish RpsQ from the other known Rps genes on chromosome 3. Identical polymerase chain reaction amplification products were produced for cultivars Qichadou 1 (RpsQ) and Ludou 4 (Rps9). All other cultivars carrying Rps genes on chromosome 3 produced different PCR products, which all lacked a 144-bp fragment present in Qichadou 1 and Ludou 4. The phenotypes of the analyzed cultivars combined with the physical position of the PRR resistance locus, candidate gene analyses, and the candidate gene marker test revealed RpsQ and Rps9 are likely the same gene, and confer resistance to P. sojae.




Similar content being viewed by others
References
Abney TS, Melgar JC, Richards TL, Scott DH, Grogan J, Young J (1997) New races of Phytophthora sojae with Rps1–d virulence. Plant Dis 81:653–655
Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor–like serine threonine kinases: roles in signaling and plant defense. Mol Plant Microbe In 21:507–517
Anderson TR, Buzzell RI (1992) Inheritance and linkage of the Rps7 gene for resistance to Phytophthora rot of soybean. Plant Dis 76:958–959
Bai JF et al (2002) Diversity in nucleotide binding site–leucine–rich repeat genes in cereals. Genome Res 12:1871–1884
Bernard RL, Smith PE, Kaufmann MJ, Schmitthenner AF (1957) Inheritance of resistance to Phytophthora root rot and stem rot in the soybean. Agron J 49:391
Braun DM, Walker JC (1996) Plant transmembrane receptors: new pieces in the signaling puzzle. Trends Biochem Sci 21:70–73
Buzzell RI, Anderson TR (1992) Inheritance and race reaction of a new soybean Rps1 allele. Plant Dis 76:600–601
Chen QH, Wang QY, Wang YC, Zheng XB (2004) Identification and sequencing of ribosomal DNA-ITS of Phytophthora sojae in Fujian. Acta Phytopathol Sin 34:112–116
Cregan PB et al (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
Cui L, Yin W, Tang Q, Dong S, Zheng X, Zhang Z, Wang Y (2010) Distribution, pathotypes, and metalaxyl sensitivity of Phytophthora sojae from Heilongjiang and Fujian provinces in China. Plant Dis 94:881–884
Dogimont C, Chovelon V, Pauquet J, Boualem A, Bendahmane A (2014) The Vat locus encodes for a CC-NBS-LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii-mediated virus resistance. Plant J 80:993–1004
Dorrance AE, Grünwald NJ (2009) Phytophthora sojae: Diversity among and within populations. In: Lamour K, Kamoun S (eds) Oomycete genetics and genomics: diversity, interactions, and research tools. J. Wiley & Sons, New Jersey, pp 197–212
Dorrance AE, Schmitthenner AF (2000) New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis 84:1303–1308
Dorrance AE, McClure SA, DeSilva A (2003) Pathogenetic diversity of Phytophthora sojae in Ohio soybean fields. Plant Dis 87:139–146
Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506
Fan AY, Wang XM, Fang XP, Wu XF, Zhu ZD (2009) Molecular identification of Phytophthora resistance gene in soybean cultivar Yudou 25. Acta Agron Sin 35:1844–1850
Förster H, Tyler BM, Coffey MD (1994) Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. MPMI 7:780–791
Gachomo EW, Shonukan OO, Kotchoni SO (2003) The molecular initiation and subsequent acquisition of disease resistance in plants. Afr J Biotechnol 2:26–32
Gao H, Bhattacharyya MK (2008) The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol 8:29. doi:10.1186/1471-2229-8-29
Gómez–Gómez L, Boller T (2000) FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011
Gordon SG, St Martin SK, Dorrance AE (2006) Rps8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci 46:168–173
Graham MA, Marek LF, Shoemaker RC (2002) Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 162:1961–1977
Grau CR, Dorrance AE, Bond J, Russin J (2004) Fungal diseases. In: Boerma HR, Specht JE (eds) Soybeans: improvement, production and uses, 3rd edn. Agronomy Monogr. American Soc Agron Madison WI, pp 679–763
Gunadi A (2012) Characterization of Rps8 and Rps3 resistance genes to Phytophthora sojae through genetic fine mapping and physical mapping of soybean chromosome 13. Dissertation, The Ohio State University
Haas JH, Buzzell RI (1976) New races 5 and 6 of Phytophthora megasperma var. sojae and differential reactions of soybean cultivars for races 1 and 6. Phytopathology 66:1361–1362
Heese A et al (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Pro Natl Aca Sci USA 104:12217–12222
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Jee HJ, Kim WG, Cho WD (1998) Occurrence of Phytophthora root rot on soybean (Glycine max) and identification of the causal fungus. RDA J Crop Prot Korea Repub
Kaitany RC, Hart LP, Safir GR (2001) Virulence composition of Phytophthora sojae in Michigan. Plant Dis 85:1103–1106
Kajava AV, Kobe B (2002) Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci 11:1082–1090
Kasuga T, Salimath SS, Shi J, Gijzen M, Buzzell RI, Bhattacharyya MK (1997) High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. MPMI 10:1035–1044
Kaufmann MJ, Gerdemann JW (1958) Root and stem rot of soybean caused by Phytophthora sojae n. sp. Phytopathology 48:201–208
Lee S, Mian R, McHale LK, Wang H, Wijeratne AJ, Sneller CH, Dorrance AE (2013) Novel quantitative trait loci for partial resistance to Phytophthora sojae in soybean PI 398841. Theor Appl Genet 126:1121–1132
Li XP et al (2006) Identification and functional characterization of a leucine-rich repeat receptor-like kinase gene that is involved in regulation of soybean leaf senescence. Plant Mol Biol 61:829–844
Lin F et al (2013) Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor Appl Genet 126:2177–2185
Lincoln SE, Daly MJ, Lander ES (1993) Construction of a genetic linkage map with MAPMAKER/EXP v3.0: a tutorial and reference manual. An whitehead institute technical report, Cambridge
Liu JL, Liu XL, Dai LY, Wang GL (2007) Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genom 34:765–776
Michelmore RW, Paran I, Kessell RV (1991) Identification of markers linked to disease-resistance genes by bulked segregate analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. P Natl Acad Sci USA 88:9828–9832
Morillo SA, Tax FE (2006) Functional analysis of receptor-like kinases in monocots and dicots. Curr Opin Plant Biol 9:460–469
Mueller EH, Athow KL, Laviolette FA (1978) Inheritance of resistance to four physiologic races of Phytophthora megasperma var. sojae. Phytopathology 68:1318–1322
Ortega MA, Dorrance AE (2011) Microsporogenesis of Rps8/rps8 heterozygous soybean lines. Euphytica 181:77–88
Parlevliet JE (2002) Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124:147–156
Pathan MS, Sleper DA (2008) Advances in soybean breeding. In: Stacey G (ed) Genetics and genomics of soybean. Springer, New York (NY), pp 113–134
Ping J et al (2015) Identification and molecular mapping of Rps11, a novel gene conferring resistance to Phytophthora sojae in soybean. Theor Appl Genet 1–7
Qiu BX, Sleper DA, Arelli AR (1997) Genetic and molecular characterization of resistance to Heterodera glycines race isolates 1, 3, and 5 in Peking. Euphytica 96:225–231
SaghaiMaroof MA, Tucker DM, Tolin SA (2008) Genomics of viral–soybean interactions. In: Stacey G (ed) Genetics and genomics of soybean. Springer Science and Business Media, New York, pp 293–319
Sandhu D, Gao H, Cianzio S, Bhattacharyya MK (2004) Deletion of a disease resistance nucleotide-binding-site leucine-rich-repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics 168:2157–2167
Schmitthenner AF (1985) Problems and progress in control of Phytophthora root rot of soybean. Plant Dis 69:362–368
Schmitthenner AF (1999) Phytophthora rot of soybean. In: Hartman GL, Sinclair JB, Rupe JC (eds) Compendium of soybean diseases, 4th edn. The American Phytopathological Society Press (APS), St Paul, Minnesota, pp 39–42
Shen CY, Su YC (1991) Discovery and preliminary studies of Phytophthora megasperma on soybean in China. Acta Phytopathol Sin 21:298
Sidhu GS, Khush GS (1978) Dominance reversal of a bacterial blight resistance gene in some rice cultivars. Phytopathology 68:461–463
Song WY et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Song Q et al (2010) Abundance of SSR motifs and development of candidate polymorphic SSR markers (BArCSOYSSr_1.0) in soybean. Crop Sci 50:1950–1960
Sugimoto T et al (2011) Genetic analysis and identification of DNA markers linked to a novel Phytophthora sojae resistance gene in the Japanese soybean cultivar Waseshiroge. Euphytica 182:133–145
Sugimoto T et al (2012) Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed Sci 61:511–522
Sun S, Wu XL, Zhao JM, Wang YC, Tang QH, Yu DY, Gai JY, Xing H (2011) Characterization and mapping of RpsYu25, a novel resistance gene to Phytophthora sojae. Plant Breed 130:139–143
Sun J, Li L, Zhao J, Huang J, Yan Q, Xing H, Guo N (2014) Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean [Glycine max (L.) Merr.]. Theor Appl Genet 127:913–919
Tyler BM (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol 8:1–8
Walters D (2009) Disease control kin crops: biological and environmentally-friendly approaches. Blackwell Publishing, Chichester
Wang Q et al (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23:2064–2086
Weng C, Yu K, Anderson TR, Poysa V (2001) Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J Hered 92:442–446
Wrather JA, Koenning SR (2006) Estimates of disease effects on soybean yields in the United States 2003 to 2005. J Nematol 38:173
Wu XL et al (2011) Identification, genetic analysis and mapping of resistance to Phytophthora sojae of Pm28 in soybean. Agr Sci China 10:1506–1511
Xia CJ, Zhang JQ, Wang XM, Wu XF, Liu ZX, Zhu ZD (2011a) Analyses of resistance genes to Phytophthora root rot in soybean germplasm. Chin J Oil Crop Sci 33:396–401
Xia CJ, Zhang JQ, Wang XM, Liu ZX, Zhu ZD (2011b) Analysis of genes resistance to Phytophthora root rot in soybean germplasm imported from America. Acta Agron Sin 37:1167–1174
Xiang Y, Cao Y, Xu C, Li X, Wang S (2006) Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor Appl Genet 113(7):1347–1355
Xue AG, Marchand G, Chen Y, Zhang S, Cober ER, Tenuta A (2015) Races of Phytophthora sojae in Ontario, Canada, 2010–2012. Can J Plant Pathol 37:376–383
Yao HY, Wang XM, Wu XF, Xiao YN, Zhu ZD (2010) Molecular mapping of Phytophthora resistance gene in soybean cultivar Zaoshu18. J Plant Genet Resour 11:213–217
Yu AL et al (2010) Genetic analysis and SSR mapping of gene resistance to Phytophthora sojae race 1 in soybean cv Suinong 10. Chin J Oil Crop Sci 32:462–466
Zhang S et al (2010) Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis 94:87–91
Zhang J, Xia C, Duan C, Sun S, Wang X, Wu X, Zhu Z (2013a) Identification and candidate gene analysis of a novel Phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PloS One 8:e69799
Zhang J, Xia C, Wang X, Duan C, Sun S, Wu X, Zhu Z (2013b) Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor Appl Genet 126:1555–1561
Zhong C, Li YP, Sun SL, Liu ZX, Qiu LJ, Zhu ZD (2015) Identification of resistance and tolerance to Phytophthora sojae in wild soybean germplasm. J Plant Genet Resour 16:684–690
Zhu ZD, Wang HB, Wang XM, Chang RZ, Wu XF (2003) Distribution and virulence diversity of Phytophthora sojae in China. Sci Agric Sin 36:793–799
Zhu ZD, Huo YL, Wang XM, Huang JB, Wu XF (2006) Screening for resistance sources to Phytophthora root rot in soybean. J Plant Genet Resour 7:24–30
Zhu ZD, Huo YL, Wang XM, Huang JB, Wu XF (2007) Molecular identification of a novel Phytophthora resistance gene in soybean. Acta Agron Sin 33:154–157
Acknowledgements
The work was supported by the Special Fund for Agroscientific Research in the Public Interest (201303018), the Program of Protection of Crop Germplasm Resources (2015NWB030-14) from the Ministry of Agriculture of the People’s Republic of China, and the Scientific Innovation Program of Chinese Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments were performed in accordance with all relevant Chinese laws.
Additional information
Communicated by Henry T. Nguyen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Sun, S., Zhong, C. et al. Genetic mapping and development of co-segregating markers of RpsQ, which provides resistance to Phytophthora sojae in soybean. Theor Appl Genet 130, 1223–1233 (2017). https://doi.org/10.1007/s00122-017-2883-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2883-7