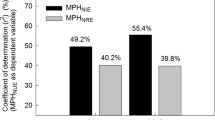
Abstract
Key message
Exploring and understanding the genetic basis of cob biomass in relation to grain yield under varying nitrogen management regimes will help breeders to develop dual-purpose maize.
Abstract
With rising energy demands and costs for fossil fuels, alternative energy from renewable sources such as maize cobs will become competitive. Maize cobs have beneficial characteristics for utilization as feedstock including compact tissue, high cellulose content, and low ash and nitrogen content. Nitrogen is quantitatively the most important nutrient for plant growth. However, the influence of nitrogen fertilization on maize cob production is unclear. In this study, quantitative trait loci (QTL) have been analyzed for cob morphological traits such as cob weight, volume, length, diameter and cob tissue density, and grain yield under normal and low nitrogen regimes. 213 doubled-haploid lines of the intermated B73 × Mo17 (IBM) Syn10 population have been resequenced for 8575 bins, based on SNP markers. A total of 138 QTL were found for six traits across six trials using composite interval mapping with ten cofactors and empirical comparison-wise thresholds (P = 0.001). Despite moderate to high repeatabilities across trials, few QTL were consistent across trials and overall levels of explained phenotypic variance were lower than expected some of the cob trait × trial combinations (R 2 = 7.3–43.1 %). Variation for cob traits was less affected by nitrogen conditions than by grain yield. Thus, the economics of cob usage under low nitrogen regimes is promising.



Similar content being viewed by others
Notes
© 2012 The MathWorks, Inc. MATLAB and Simulink are registered trademarks of The MathWorks, Inc. See www.mathworks.com/trademarks for a list of additional trademarks. Other product or brand names may be trademarks or registered trademarks of their respective holders.
References
Adams KR, Muenchrath DA, Schwindt DM (1999) Moisture effects on the morphology of ears, cobs and kernels of a south-western U.S. maize (Zea maysL.) cultivar, and implications for the interpretation of archaeological maize. J Archaeol Sci 26( 5):483–496
Andorf CM, Lawrence CJ, Harper LC, Schaeffer ML, Campbell DA, Sen TZ (2010) The Locus Lookup tool at MaizeGDB: identification of genomic regions in maize by integrating sequence information with physical and genetic maps. Bioinformatics 26:434–436
Austin DF, Lee M, Veldboom LR, Hallauer AR (2000) Genetic mapping in maize with hybrid progeny across testers and generations: grain yield and grain moisture. Crop Sci 40:30–39
Basten CJ, Weir BS, Zeng ZB (2003) QTL cartographer version 1.17. North Carolina State University, Raleigh, NC, USA
Bennewitz J, Reinsch N, Kalm E (2002) Improved confidence intervals in quantitative trait loci mapping by permutation bootstrapping. Genetics 160(4):1673–1686
Bohning K (2000) US patent 6084163, Inbred corn line BE4547
Cooper M, Delacy IH (1994) Relationships among analytical methods used to study genotypic variation and genotype-by-environment interaction in plant breeding multi-environment experiments. Theor Appl Genet 88:561–572
Crossett A, Lauter Nick, Love Tanzy M (2010) An empirical method for establishing positional confidence intervals tailored for composite interval mapping of QTL. PLoS One 5(2):e9039
Galinat WC (1975) The evolutionary emergence of maize. Bull Torrey Bot Club 102(6):313–324
Ganal MW, Durstewitz G, Polley A, Bérard Al, Buckler ES, Charcosset A, Clarke JD, Graner E-M, Hansen M, Joets J, Le Paslier M-C, McMullen MD, Montalent P, Rose M, SchÃn C-C, Sun Q, Walter H, Martin OC, Falque M (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One 6(12):e28334
George E, Marschner H, Jakobsen I (1995) Role of arbuscular mycorrhizal fungi in uptake of phosphorus and nitrogen from soil. Crit Rev Biotechnol 15(3–4):257–270
Haldane JBS (1919) The combination of linkage values and the calculation of distance between the loci of linked factors. J Genet 8:299–309
Henderson CR (1975) Best linear unbiased prediction under a selection model. Biometrics 31:423–447
Holland JB (2006) Estimating genotypic correlations and their standard errors using multivariate restricted maximum likelihood estimation with SAS Proc MIXED. Crop Sci 46(2):642–654
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Hussain T, Tausend P, Graham G, Ho J (2007) Registration of IBM2 SYN10 doubled haploid mapping population of maize. J Plant Regist 1:81–81
Hyne V, Kearsey M, Pike D, Snape J (1995) QTL analysis: unreliability and bias in estimation procedures. Mol Breed 1(3):273–282
Jansen C, Lübberstedt T (2012) Turning maize cobs into a valuable feedstock. Bio Energy Res 5:20–31
Jansen C, de Leon N, Lauter N, Hirsch C, Ruff L, Lübberstedt T (2013) Genetic and morphometric analysis of cob architecture and biomass-related traits in the intermated B73 × Mo17 recombinant inbred lines of maize. Bioenerg Res 6:903–916
Knapp SJ, Bridges WC (1990) Using molecular markers to estimate quantitative trait locus parameters: power and genetic variances for unreplicated and replicated progeny. Genetics 126:769–777
Lauter N, Moscou MJ, Habiger J, Moose SP (2008) Quantitative genetic dissection of shoot architecture traits in maize: towards a functional genomics approach. Plant Genome J 1(2):99–110
Lee M, Sharopova N, Beavis WD, Grant D, Maria Katt M, Blair D, Hallauer A (2002) Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol Biol 48(5–6):453–461
Loesch PJ, Stark CF, Zuber MS (1976) Effects of plant density on the quality of cobs used for corn cob pipes. Alliance Crop Soil Environ Sci Soc 16(5):706–709
MaizeGDB http://www.maizegdb.org. Accessed 6 July 2012
Melchinger AE, Utz HF, Schön CC (1998) Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149(1):383–403
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
Murdy WH (1960) The strengthening system in the stem of maize. Ann Mo Bot Gard 47(3):205–226
Openshaw S, Frascaroli E (1997) QTL detection and marker assisted selection for complex traits in maize. In: 52nd annual corn and sorghum industry research conference. ASTA, Washington, DC, pp 44–53
Power J, Schepers J (1989) Nitrate contamination of groundwater in North America. Agric Ecosyst Environ 26(3–4):165–187
Van Oijen JW (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84(7):803–811
Van Ooijen JW (1999) LOD signifcance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83:613–624
Vattikonda M (2000) US patent 6137038, Inbred corn line SM4603
Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, Schindler DW et al (1997) Human alteration of the global nitrogen cycle: sources and consequences. Ecol Appl 7(3):737–750
Vollbrecht E, Springer PS, Goh L, Buckler ES, Martienssen R (2005) Architecture of floral branch systems in maize and related grasses. Nature 436(25):1119–1126
Zhang L, Wang S, Li H, Deng Q, Zheng A, Li S, Li P, Li Z, Wang J (2010) Effects of missing marker and segregation distortion on QTL mapping in F2 populations. Theor Appl Genet 121:1071–1082
Acknowledgments
The authors would like to acknowledge Pioneer Hi-Bred for growing the IBMsyn10 population at Marion, IA in 2010 and for providing phenotyping capacities at that location. The Authors also would like to thank USDA_s National Institute of Food and Agriculture (project number: IOW05180) for funding this work. Constantin Jansen and Pedro J. Gonzalez-Portilla were supported by the Interdepartmental Genetics Graduate Program as well as RF Baker Center for Plant Breeding at Iowa State University. Yongzhong Zhang and Hongjun Liu were the visiting student at ISU, supported by China Scholarship Council.
Conflict of interest
The authors declare that no conflict of interest exists.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by J. Yan.
C. Jansen, Y. Zhang, H. Liu and P. J. Gonzalez-Portilla contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jansen, C., Zhang, Y., Liu, H. et al. Genetic and agronomic assessment of cob traits in corn under low and normal nitrogen management conditions. Theor Appl Genet 128, 1231–1242 (2015). https://doi.org/10.1007/s00122-015-2486-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2486-0