Abstract
Powdery mildew (PM) is a very important disease of cucumber (Cucumis sativus L.). Resistant cultivars have been deployed in production for a long time, but the genetic mechanisms of PM resistance in cucumber are not well understood. A 3-year QTL mapping study of PM resistance was conducted with 132 F2:3 families derived from two cucumber inbred lines WI 2757 (resistant) and True Lemon (susceptible). A genetic map covering 610.4 cM in seven linkage groups was developed with 240 SSR marker loci. Multiple QTL mapping analysis of molecular marker data and disease index of the hypocotyl, cotyledon and true leaf for responses to PM inoculation identified six genomic regions in four chromosomes harboring QTL for PM resistance in WI 2757. Among the six QTL, pm1.1 and pm1.2 in chromosome 1 conferred leaf resistance. Minor QTL pm3.1 (chromosome 3) and pm4.1 (chromosome 4) contributed to disease susceptibility. The two major QTL, pm5.1 and pm5.2 were located in an interval of ~40 cM in chromosome 5 with each explaining 21.0–74.5 % phenotypic variations. Data presented herein support two recessively inherited, linked major QTL in chromosome 5 plus minor QTL in other chromosomes that control the PM resistance in WI 2757. The QTL pm5.2 for hypocotyl resistance plays the most important role in host resistance. Multiple observations in the same year revealed the importance of scoring time in the detection of PM resistance QTL. Results of this study provided new insights into phenotypic and genetic mechanisms of powdery mildew resistance in cucumber.



Similar content being viewed by others
References
Arends D, Prins P, Jansen RC, Broman KW (2010) R/qtl: high-throughput multiple QTL mapping. Bioinformat Appl Note 2:2990–2992
Barnes WC (1961) Multiple disease resistant cucumbers. Proc Amer Soc Hort Sci 77:417–423
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTLmapping in experimental crosses. Bioinformatics 19:889–890
Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD, Huang S, Weng Y (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics 11:569
Darvasi A, Soller M (1995) Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141:1199–1207
de Ruiter W, Hofstede R, de Vries J, van den Heuvel H (2008) Combining QTL for resistance to CYSDV and powdery mildew in a single cucumber line. In: Proc 9th EUCARPIA meeting on genetics and breeding of Cucurbitaceae (Pitrat M, ed), INRA, Avignon (France), May 21–24, 181–188
Fanourakis N, Simon PW (1987) Analysis of genetic linkage in cucumber. J Hered 78:238–242
Fujieda K, Akiya R (1962) Genetic study of powdery mildew resistance and spine color on fruit in cucumber. J Jap Soc Hort Sci 31:30–32
Hofstede R, de Ruiter W, de Vries RJ, van den Heuvel H (2008) Disease resistant cucumber plants. US Patent (# US 2008/0307540 A1)
Kooistra E (1968) Powdery mildew resistance in cucumber. Euphytica 17:236–242
Kooistra E (1971) Inheritance of fruit flesh and skin colors in powdery milew resistant cucumbers (Cucumis sativus L.). Euphytica 20:521–523
Li Y, Yang L, Pathak M, Li D, He X, Weng Y (2011) Fine genetic mapping of cp: a recessive gene for compact (dwarf) plant architecture in cucumber, Cucumis sativus L. Theor Appl Genet 123:973–983
Liu LZ, Yuan XJ, Cai R, Pan JS, He HL, Yuan LH, Guan Y, Zhu LH (2008) Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. J Phytopathol 156:691–697
Mather K, Jinks JL (1971) Biometrical genetics. Chapman and Hall, London
Miao H, Zhang SP, Wang XW, Zhang ZH, Li M, Mu SQ et al (2011) A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 182:167–176
Perez-Garcia A, Romero D, Fernandez-Ortuno D, Lopez-Ruiz F, De Vicente A, Tores JA (2009) The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol Plant Pathol 10:153–160
Peterson CE, Williams PH, Palmer M, Louward P (1982) Wisconsin 2757 Cucumber. HortSci 17:268
Ren Y, Zhang Z, Liu J, Staub JE, Han Y, Cheng Z, Li X et al (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 4:e5795
Sakata Y, Kubo N, Morishita M, Kitadani E, Sugiyama M, Hirai M (2006) QTL analysis of powdery mildew resistance in cucumber. Theor Appl Genet 112:243–250
Shanmugasundaram S, Williams PH, Peterson CE (1971) Inheritance of resistance to powery mildew in cucumber. Phytopathol 61:1218–1221
Sitterly WP (1978) Powdery mildew of cucurbits. In: Spencer DM (ed) The powdery mildews. Academic Press, London, pp 359–379
Smith PG (1948) Powdery mildew resistance in cucumber. Phytopathol 38:1027–1028
Stuber CW, Edwards MD, Wendel JF (1987) Molecular marker-facilitated investigations of quantitative trait loci in maize: II. Factors influencing yield and its component traits. Crop Sci 27:639–648
Vakalounakis DJ (1992) Heart leaf, a recessive leaf shape marker in cucumber linkage with disease resistance and other traits. J Hered 83:217–221
Vliet GJAV, Meijsing WD (1977) Relation in inheritance of resistance to Pseudoperonospora cubensis Rost and Sphaerotheca fuliginea Poll in cucumber (Cucumis sativus L). Euphytica 26:793–796
Walters SA, Shetty NV, Wehner TC (2001) Segregation and linkage of several genes in cucumber. J Am Soc Hortic Sci 126:442–450
Yang L, Koo DH, Li Y, Zhang X, Luan F, Havey MJ, Jiang J, Weng Y (2012) Chromosome rearrangements during domestication of cucumber as revealed from high-density genetic mapping and draft genome assembly. Plant J 71:895–906
Zeng ZB, Kao CH, Basten CJ (1999) Estimating the genetic architecture of quantitative traits. Genet Res 74:279–289
Zhang HY, Wang ZG, Mao AJ, Zhang F, Wang YJ, Xu Y (2008) SSR markers linked to the resistant gene of cucumber powdery mildew. Acta Agri Boreali-Sinica 23:77–80
Zhang SP, Liu MM, Miao H, Zhang SQ, Yang YH, Xie BY, Gu XF (2011) QTL mapping of resistance genes to powdery mildew in cucumber. Scientia Agri Sinica 44:3584–3593
Zitter TA, Hopkins DL, Thomas CE (1996) Compendium of cucurbits diseases. APS Press, Saint Paul
Acknowledgments
The authors thank Linda Crubaugh for technical assistance and two anonymous reviewers for critical reading and valuable suggestions to improve an early version of the manuscript. XH was supported by the Guangdong Academy of Agricultural Sciences, China. YL was supported by China Scholarship Council. MP was supported by a training grant from Punjab Agricultural University, Ludhiana, India. SP was supported by a training grant from the Indian Council of Agricultural Research, New Delhi, India. The authors greatly appreciate the support of these sponsors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Paran.
X. He and Y. Li contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
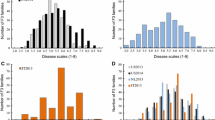
Fig. S1 Frequency distribution of DI for hypocotyl (A), cotyledons (B) and true leaf (C) PM resistances in EXPT1 (2010), EXPT2 (2011) and second observation of EXPT3. DI means for parental lines, F1 and controls are shown in the table.
Fig. S2 Frequency distribution of DI for hypocotyl (HY, A), cotyledons (CL, B) and true leaf (TL, C) PM resistance from three observations of EXPT3 (EXPT3-1, EXPT3-2 and EXPT3-3). Data of the three observations were collected at four-day intervals.
Fig. S3 A SSR-based linkage map developed with 132 WI 2757 × True Lemon F2 plants. Numbers on top of the map are chromosomes numbers. Cumulative map distance (cM) is shown to the left of each linkage group and marker designation is on the right. The gene pm-h at 83.3 cM in Chr5 is for hypocotyl PM resistance in WI 2757 based on EXPT2 data.
Fig. S4 LOD curves of QTL for PM resistance based on phenotypic data from three observations in EXPT3 demonstrating effects of scoring time on QTL detection. A global view of map locations of ten QTL is presented in A. LOD curves of seven QTLs mapped in chromosome 5 are shown in B. LOD profiles were based on simple interval mapping, which differed slightly from MQM profiles shown in Table 2 for some QTL. The dashed line is LOD threshold (3.5) based on 10,000 permuted samples.
Table S1. Major statistics of a linkage map developed with 132 F2 plants of WI 2757 × True Lemon.
Table S2. Information of 240 markers placed on the WI2757 × True Lemon F2 genetic map. Marker loci are arranged by increasing order of map locations in each linkage group (LG). The physical location of each marker in the Gy14 scaffold assembly is also shown.
Rights and permissions
About this article
Cite this article
He, X., Li, Y., Pandey, S. et al. QTL mapping of powdery mildew resistance in WI 2757 cucumber (Cucumis sativus L.). Theor Appl Genet 126, 2149–2161 (2013). https://doi.org/10.1007/s00122-013-2125-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2125-6