Abstract
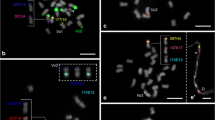
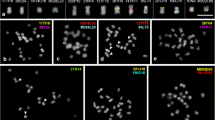
The common bean (Phaseolus vulgaris) and lima bean (P. lunatus) are among the most important legumes in terms of direct human consumption. The present work establishes a comparative cytogenetic map of P. lunatus, using previously mapped markers from P. vulgaris, in association with analyses of heterochromatin distribution using the fluorochromes chromomycin A3 (CMA) and 4′,6-diamidino-2-phenylindole (DAPI) and localization of the 5S and 45S ribosomal DNA (rDNA) probes. Seven BACs selected from different common bean chromosomes demonstrated a repetitive pericentromeric pattern corresponding to the heterochromatic regions revealed by CMA/DAPI and could not be mapped. The subtelomeric repetitive pattern observed for BAC 63H6 in most of the chromosome ends of common bean was not detected in lima bean, indicating lack of conservation of this subtelomeric repeat. All chromosomes could be identified and 16 single-copy clones were mapped. These results showed a significant conservation of synteny between species, although change in centromere position suggested the occurrence of pericentric inversions on chromosomes 2, 9 and 10. The low number of structural rearrangements reflects the karyotypic stability of the genus.



Similar content being viewed by others
References
Adam-Blondon A, Sévignac M, Dron M, Bannerot H (1994) A genetic map of common bean to localize specific resistance genes against anthracnose. Genome 37:915–924
Almeida C, Pedrosa-Harand A (2010) Contrasting rDNA evolution in lima bean (Phaseolus lunatus L.) and common bean (P. vulgaris L., Fabaceae). Cytogenet Genome Res 132:212–217
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Broughton WJ, Hernández G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.): model food legumes. Plant Soil 252:55–128
Cabral JS, Felix LP, Guerra M (2006) Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae). Genet Mol Biol 29:659–664
Carvalho R, Filho WSS, Brasileiro-Vidal AC, Guerra M (2005) The relationship among lemons, limes and citron: a chromosomal comparison. Cytogenetic Genome Res 109:276–282
Chacón SMI, Pickersgill B, Debouck DG (2005) Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races. Theor Appl Genet 110:432–444
David P, Chen NWG, Pedrosa-Harand A et al (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151:1048–1065
Deumling B, Greilhuber J (1982) Characterization of heterochromatin in different species of the Scilla siberica group (Liliaceae) by in situ hybridization of satellite DNAs and fluorochrome banding. Chromosoma 84:535–555
Doganlar S, Frary A, Daunay M-C, Lester RN, Tanksley SD (2002) A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161:1697–1711
Dubcovsky J, Dvorak J (1995) Ribossomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140:1367–1377
Ferrier-Cana E, Geffroy V, Macadré C et al (2003) Characterization of expressed NBS-LRR resistance gene candidates from common bean. Theor Appl Genet 106:251–261
Fonsêca A, Ferreira J, Santos TRB et al (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosome Res 8:487–502
Freyre R, Skroch PW, Geffroy V et al (1998) Towards an integrated linkage map of common bean 4: development of a core linkage map and alignment of RFLP maps. Theor Appl Genet 97:847–856
Geffroy V, Sévignac M, Billiant P, Dron M, Langin T (2008) Resistance to Colletotrichum lindemuthianum in Phaseolus vulgaris: a case study for mapping two independent genes. Theor Appl Genet 116:407–415
Geffroy V, Macadré C, David P et al (2009) Molecular analysis of a large subtelomeric NBS-LRR family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181:405–419
Han Y, Zhang Z, Liu C et al (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci USA 106:14937–14941
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA:DNA in situ hybridization preparations. Trends Genet 8:372–373
Hizume M, Ohgiku A, Tanaka A (1989) Chromosome banding in the genus Pinus II. Interespecific variation on fluorescent banding patterns in P. densiflora e P. thunbergii. Bot Mag 102:25–36
Iovene M, Wielgus SM, Simom PH, Buell CR, Jiang J (2008) Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 180:1307–1317
Iovene M, Cavagnaro PF, Senalik D, Buell CR, Jiang J, Simon PW (2011) Comparative FISH mapping of Daucus species (Apiaceae family). Chromosome Res 19:493–506
Kami J, Poncet V, Geffroy V, Gepts P (2006) Development of four phylogenetically-arrayed BAC libraries and sequence of the APA locus in Phaseolus vulgaris. Theor Appl Genet 112:987–998
Koumbaris GL, Bass HW (2003) A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propiquum L.) BAC clones. Plant J 35:647–659
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA 103:5224–5229
Ma L, Vu GTH, Schubert V et al (2010) Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res 18:841–850
Mandáková T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20:2559–2570
Men AE, Meksem K, Kassem MA et al (2001) A bacterial chromosome library of Lotus japonicus constructed in an Agrobacterium tumefaciens-transformable vector. MPMI 14:422–425
Mercado-Ruaro P, Delgado-Salinas A (1996) Karyological studies in several Mexican species of Phaseolus L. and Vigna savi (Phaseolinae, Fabaceae). In: Pickergill B, Lock JM (eds) Advances in legumes systematics 8: legumes of economic importance. Royal Botanic Gardens, Kew, pp 83–87
Mercado-Ruaro P, Delgado-Salinas A (1998) Karyotypic studies on species of Phaseolus (Fabaceae: Phaseolinae). Am J Bot 85:1–9
Moscone EA, Klein F, Lambrou M, Fuchs J, Schweizer D (1999) Quantitative karyotyping and dual-color FISH mapping of 5S and 18S–25S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 42:1224–1233
Nodari RO, Tsai SM, Gilbertson RL, Gepts P (1993) Towards an integrated linkage map of common bean: II development of an RFLP-based linkage map. Theor Appl Genet 85:513–520
Oliveira AP, Alves EU, Alves AU et al (2004) Produção de feijão-fava em função do uso de doses de fósforo em um Neossolo Regolítico. Hortic Bras 22:543–546
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Pedrosa A, Vallejos CE, Bachmair A, Schweizer D (2003) Integration of common bean (Phaseolus vulgaris L.) linkage and chromosomal maps. Theor Appl Genet 106:205–212
Pedrosa-Harand A, Porch T, Gepts P (2008) Standard nomenclature for common bean chromosomes and linkage groups. Annu Rep Bean Improv Coop 51:106–107
Pedrosa-Harand A, Kami J, Gepts P, Geffroy V, Schweizer D (2009) Cytogenetic mapping of common bean chromosomes reveals a less compartmentalised small-genome plant species. Chromosome Res 17:405–417
Raskina O, Belyayev A, Nevo E (2004) Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc Natl Acad Sci USA 101:14818–14823
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27:207–2016
Serrano–Serrano ML, Hernández-Torres J, Castillo-Villamizar G, Debouck DG, Sánchez MI (2010) Gene pools in wild Lima bean (Phaseolus lunatus L.) from the Americas: evidences for an Andean origin and past migrations. Mol Phylogenet Evol 54:76–87
Silva SC, Martins MIG, Santos RC et al (2010) Karyological features and banding patterns in Arachis species belonging to the Heteranthae section. Plant Syst Evol 285:201–207
Souza MGC, Benko-Iseppon AM (2004) Cytogenetics and chromosome banding patterns in Caesalpinioideae and Papilionioideae species of Pará, Amazonas, Brazil. Bot J Linn Soc 144:181–191
Tang X, Szinay D, Lang C et al (2008) Cross-species bacterial artificial chromosome–fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements. Genetics 180:1319–1328
Topp CN, Okagaki RJ, Melo JR, Kynast RG, Phillips RL, Dawe RK (2009) Identification of a maize neocentromere in an oat-maize addition line. Cytogenet Genome Res 124:228–238
Vallejos CE, Sakiyama NS, Chase CD (1992) A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131:733–740
Wanzenböck E-M, Schöfer C, Schweizer D, Bachmair A (1997) Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J 11:1007–1016
Wu C–C, Nimmakayala P, Santos FA et al (2004) Construction and characterization of a soybean bacterial artificial chromosome library and use of multiple complementary libraries for genome physical mapping. Theor Appl Genet 109:1041–1050
Yang J, Wang Q, Deng D et al (2003) Construction and characterization of a bacterial artificial chromosome library of maize inbred line 77Ht2. Plant Mol Biol Rep 21:159–169
Zheng J, Nakata M, Uchiyama H, Morikawa H, Tanaka R (1991) Giemsa C-banding patterns in several species of Phaseolus L. and Vigna savi, Fabaceae. Cytologia 56:459–466
Acknowledgments
We thank Heloisa Torres (EMBRAPA Arroz e Feijão) for the seeds, Dr. Paul Gepts (University of California) for the BAC clones and Valérie Geffroy (Université Paris Sud) for the B61 bacteriophage. C. A. was supported by a grant from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil. A. F. and K. G. B. dos S. were supported by grants from Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE), Brazil. M. M. was supported by the Gregor Mendel Institute of Molecular Plant Biology (GMI), Austria. The work was supported by CNPq, Brazil.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Friebe.
Rights and permissions
About this article
Cite this article
Bonifácio, E.M., Fonsêca, A., Almeida, C. et al. Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor Appl Genet 124, 1513–1520 (2012). https://doi.org/10.1007/s00122-012-1806-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1806-x