Abstract
Objective
The molecular heterogeneity of prostate cancer (PCa) gives rise to distinct tumor subclasses based on epigenetic modification and gene expression signatures. Identification of clinically actionable molecular subtypes of PCa is key to improving patient outcome, and the balance between specific pathways may influence PCa outcome. It is also urgent to identify progression-related markers through cytosine-guanine (CpG) methylation in predicting metastasis for patients with PCa.
Methods
We performed bioinformatics analysis of transcriptomic, and clinical data in an integrated cohort of 551 prostate samples. The datasets included retrospective The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) cohorts. Two algorithms, Least Absolute Shrinkage and Selector Operation and Support Vector Machine-Recursive Feature Elimination, were used to select significant CpGs.
Results
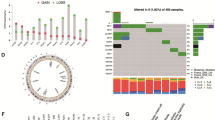
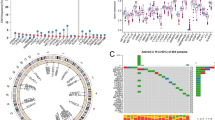
We found that PCa progression is more likely to occur after the third year through conditional survival (CS) analysis, and prostate-specific antigen (PSA) was a better predictor of Progression-free survival (PFS) than Gleason score (GS). Our study first demonstrated that PCa tumors have distinct expression profiles based on the expression of genes involved in androgen receptor (AR) and PI3K-AKT, which influence disease outcome. Our results also indicated that there are multiple phenotypes relevant to the AR-PI3K axis in PCa, where tumors with mixed phenotype may be more aggressive or have worse outcome than quiescent phenotype. In terms of epigenetics, we obtained CpG sites and their corresponding genes which have a good predictive value of PFS. However, various evidences showed that the predictive value of CpGs corresponding genes was much lower than GpG sites in Overall survival (OS) and PFS.
Conclusions
PCa classification specific to AR and PI3K pathways provides novel biological insight into previously established PCa subtypes and may help develop personalized therapies. Our results support the potential clinical utility of DNA methylation signatures to distinguish tumor metastasis and to predict prognosis and outcomes.








Similar content being viewed by others
Availability of data and materials
All data obtained and/or analyzed in this study were available from the corresponding authors in a reasonable request.
Abbreviations
- PCa:
-
Prostate cancer
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene Expression Omnibus
- CpG:
-
Cytosine-guanine
- CS:
-
Conditional survival
- PSA:
-
Prostate-specific antigen
- GS:
-
Gleason score
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- AR:
-
Androgen receptor
- CRPC:
-
Castration-resistant prostate cancer
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- GSCA:
-
Gene Set Cancer Analysis
- GDSC:
-
Genomics of Drug Sensitivity in Cancer
- LASSO:
-
Least Absolute Shrinkage and Selector Operation
- SVM-RFE:
-
Support Vector Machine-Recursive Feature Elimination
- EMT:
-
Epithelial-mesenchymal transition
References
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30
Lo UG, Lee CF, Lee MS, Hsieh JT (2017) The role and mechanism of epithelial-to-mesenchymal transition in prostate cancer progression. Int J Mol Sci 18(10):E2079
Hayes JH, Barry MJ (2014) Screening for prostate cancer with the prostate-specific antigen test: a review of current evidence. JAMA 311(11):1143–1149
Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM (2012) Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med 4:127
Damodaran S, Kyriakopoulos CE, Jarrard DF (2017) Newly diagnosed metastatic prostate cancer: has the paradigm changed? Urol Clin North Am 44(4):611–621
Waltering KK, Helenius MA, Sahu B, Manni V, Linja MJ, Jänne OA et al (2009) Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Can Res 69:8141–8149
Zoran C, Frédéric RS (2014) Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev 33:413–427
Mateo J, Smith A, Ong M, de Bono JS (2014) Novel drugs targeting the androgen receptor pathway in prostate cancer. Cancer Metastasis Rev 33(2–3):567–579
Lin Y, Lu Z, Kokontis J, Xiang J (2013) Androgen receptor primes prostate cancer cells to apoptosis through down-regulation of basal p21 expression. Biochem Biophys Res Commun 430:289–293
Cunha GR, Ricke W, Thomson A, Marker PC, Risbridger G, Hayward SW et al (2004) Hormonal, cellular, and molecular regulation of normal and neoplastic prostatic development. J Steroid Biochem Mol Biol 92:221–236
Attard G, Cooper CS, De Bono JS (2009) Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell 16(8):458–462
Manning BD, Cantley LC (2007) AKT/PKB signaling: navigating downstream. Cell 129:1261–1274
Mediwala SN, Sun H, Szafran AT, Hartig SM, Sonpavde G, Hayes TG et al (2013) The activity of the androgen receptor variant AR-V7 is regulated by FOXO1 in a PTEN-PI3K-AKTdependent way. Prostate 73:267–277
Trotta AP, Need EF, Selth LA, Chopra S, Pinnock CB, Leach DA et al (2013) Knockdown of the co-chaperone SGTA results in the suppression of androgen and PI3K/AKT signaling and inhibition of prostate cancer cell proliferation. Int J Cancer 133(12):2812–2823
Gerhauser C, Favero F, Risch T, Weischenfeldt J et al (2018) Molecular evolution of early-onset prostate cancer identifes molecular risk markers and clinical trajectories. Cancer Cell 34(6):996–1011
Abeshouse A et al (2005) Cancer Genome Atlas Research Network Te molecular taxonomy of primary prostate cancer. Cell 163:1011–1025
Bhasin JM, Lee BH, Matkin L, Ting AH et al (2015) Methylome-wide sequencing detects DNA hypermethylation distinguishing indolent from aggressive prostate Cancer. Cell Rep 13:2135–2146
Yegnasubramanian S, Kowalski J, Gonzalgo ML et al (2004) Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res 64:1975–1986
Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349:2042–2054
Skvortsova K, Masle-Farquhar E, Luu PL, Clark SJ et al (2019) DNA hypermethylation encroachment at CpG island borders in cancer is predisposed by H3K4 monomethylation patterns. Cancer Cell 35(2):297–314
Liu CJ, Hu FF, Xia MX, Guo AY et al (2018) GSCALite: a web server for gene set cancer analysis. Bioinformatics 34(21):3771–3772
Katz MHG, Hu CY, Fleming JB, Pisters PWT, Lee JE, Chang GJ (2012) Clinical calculator of conditional survival estimates for resected and unresected survivors of pancreatic cancer. Arch Surg 147(6):513–519
Kent TS, Sachs TE, Sanchez N, Vollmer CM, Callery MP (2011) Conditional survival in pancreatic cancer: better than expected. HPB 13(12):876–880
Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H et al (2009) A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Can Res 69:2305–2313
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S et al (2011) Reciprocal feedback regulation of PI3k and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19:575–586
Crumbaker M, Khoja L, Joshua AM (2017) AR signaling and the PI3K pathway in prostate cancer. Cancers (Basel) 9(4):1–15
Chau CH, Figg WD (2005) Molecular and phenotypic heterogeneity of metastatic prostate cancer. Cancer Biol Ther 4(2):166–177
Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL (2003) Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol 34:646–653
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C et al (2011) Reciprocal feedback regulation of PI3k and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19(5):575–586
Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP et al (2011) Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19(6):792–804
Bedford MT, Van Helden PD (1987) Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res 47:5274–5276
Paziewska A, Dabrowska M, Goryca K et al (2014) DNA methylation status is more reliable than gene expression at detecting cancer in prostate biopsy. Br J Cancer 111:781–789
Yegnasubramanian S, Háner MC, Zhang Y et al (2008) DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res 68:8954–8967
Fraser M, Sabelnykova VY, Yamaguchi TN, Boutros PC et al (2017) Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541(7637):359–364
Zhao SG, Chang SL, Erho N, Feng FY et al (2017) Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol 3(12):1663–1672
Acknowlegements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of China (No. 81972373) and Scientific Research Foundation for Advanced Talents, Xiang’an Hospital of Xiamen University (NO. PM201809170001).
Author information
Authors and Affiliations
Contributions
Conception and design: YZ, CS; provision of study materials: XH, HRY, XL, and HMS; collection and assembly of data: YZ, XH, HRY; data analysis and interpretation: YZ, XH, XL; manuscript writing: YZ, CS. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Y., Hu, X., Yu, H. et al. Alternations of gene expression in PI3K and AR pathways and DNA methylation features contribute to metastasis of prostate cancer. Cell. Mol. Life Sci. 79, 436 (2022). https://doi.org/10.1007/s00018-022-04456-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04456-2