Abstract
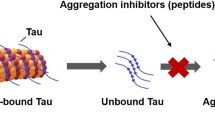
Neurofibrillary tangles of the Tau protein and plaques of the amyloid β peptide are hallmarks of Alzheimer’s disease (AD), which is characterized by the conversion of monomeric proteins/peptides into misfolded β-sheet rich fibrils. Halting the fibrillation process and disrupting the existing aggregates are key challenges for AD drug development. Previously, we performed in vitro high-throughput screening for the identification of potent inhibitors of Tau aggregation using a proxy model, a highly aggregation-prone hexapeptide fragment 306VQIVYK311 (termed PHF6) derived from Tau. Here we have characterized a hit molecule from that screen as a modulator of Tau aggregation using in vitro, in silico, and in vivo techniques. This molecule, an anthraquinone derivative named Purpurin, inhibited ~ 50% of PHF6 fibrillization in vitro at equimolar concentration and disassembled pre-formed PHF6 fibrils. In silico studies showed that Purpurin interacted with key residues of PHF6, which are responsible for maintaining its β-sheets conformation. Isothermal titration calorimetry and surface plasmon resonance experiments with PHF6 and full-length Tau (FL-Tau), respectively, indicated that Purpurin interacted with PHF6 predominantly via hydrophobic contacts and displayed a dose-dependent complexation with FL-Tau. Purpurin was non-toxic when fed to Drosophila and it significantly ameliorated the AD-related neurotoxic symptoms of transgenic flies expressing WT-FL human Tau (hTau) plausibly by inhibiting Tau accumulation and reducing Tau phosphorylation. Purpurin also reduced hTau accumulation in cell culture overexpressing hTau. Importantly, Purpurin efficiently crossed an in vitro human blood–brain barrier model. Our findings suggest that Purpurin could be a potential lead molecule for AD therapeutics.










Similar content being viewed by others
References
Maccioni RB, Munoz JP, Barbeito L (2001) The molecular bases of Alzheimer’s disease and other neurodegenerative disorders. Arch Med Res 32:367–381
Walsh DM, Selkoe DJ (2004) Deciphering the molecular basis of memory failure in Alzheimer’s disease. Neuron 44:181–193. https://doi.org/10.1016/j.neuron.2004.09.010
Bloom GS (2014) Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. https://doi.org/10.1001/jamaneurol.2013.5847
Van Cauwenberghe C, Van Broeckhoven C, Sleegers K (2016) The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med 18:421–430. https://doi.org/10.1038/gim.2015.117
Gotz J, Halliday G, Nisbet RM (2019) Molecular pathogenesis of the tauopathies. Annu Rev Pathol 14:239–261. https://doi.org/10.1146/annurev-pathmechdis-012418-012936
Mietelska-Porowska A, Wasik U, Goras M et al (2014) Tau protein modifications and interactions: their role in function and dysfunction. Int J Mol Sci 15:4671–4713. https://doi.org/10.3390/ijms15034671
Dehmelt L, Halpain S (2005) The MAP2/Tau family of microtubule-associated proteins. Genome Biol 6:204. https://doi.org/10.1186/gb-2004-6-1-204
Kadavath H, Hofele RV, Biernat J et al (2015) Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc Natl Acad Sci U S A 112:7501–7506. https://doi.org/10.1073/pnas.1504081112
Alonso A, Zaidi T, Novak M et al (2001) Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc Natl Acad Sci U S A 98:6923–6928. https://doi.org/10.1073/pnas.121119298
Barghorn S, Davies P, Mandelkow E (2004) Tau paired helical filaments from Alzheimer’s disease brain and assembled in vitro are based on beta-structure in the core domain. Biochemistry 43:1694–1703. https://doi.org/10.1021/bi0357006
Brunden KR, Ballatore C, Crowe A et al (2010) Tau-directed drug discovery for Alzheimer’s disease and related tauopathies: a focus on tau assembly inhibitors. Exp Neurol 223:304–310. https://doi.org/10.1016/j.expneurol.2009.08.031
Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT (1992) Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology 42:631–639
Kametani F, Hasegawa M (2018) Reconsideration of amyloid hypothesis and Tau hypothesis in Alzheimer’s disease. Front Neurosci 12:25. https://doi.org/10.3389/fnins.2018.00025
Irwin DJ (2016) Tauopathies as clinicopathological entities. Parkinsonism Relat Disord 22(Suppl 1):S29–S33. https://doi.org/10.1016/j.parkreldis.2015.09.020
Yamada K (2017) Extracellular tau and its potential role in the propagation of tau pathology. Front Neurosci 11:667. https://doi.org/10.3389/fnins.2017.00667
Shafiei SS, Guerrero-Muñoz MJ, Castillo-Carranza DL (2017) Tau oligomers: cytotoxicity, propagation, and mitochondrial damage. Front Aging Neurosci 9:83. https://doi.org/10.3389/fnagi.2017.00083
Ganguly P, Do TD, Larini L et al (2015) Tau assembly: the dominant role of PHF6 (VQIVYK) in microtubule binding region repeat R3. J Phys Chem B 119:4582–4593. https://doi.org/10.1021/acs.jpcb.5b00175
von Bergen M, Friedhoff P, Biernat J et al (2000) Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif [(306)VQIVYK(311)] forming beta structure. Proc Natl Acad Sci U S A 97:5129–5134
Seidler PM, Boyer DR, Rodriguez JA et al (2018) Structure-based inhibitors of tau aggregation. Nat Chem 10:170–176. https://doi.org/10.1038/nchem.2889
Fitzpatrick AWP, Falcon B, He S et al (2017) Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547:185–190. https://doi.org/10.1038/nature23002
Goux WJ, Kopplin L, Nguyen AD et al (2004) The formation of straight and twisted filaments from short tau peptides. J Biol Chem 279:26868–26875. https://doi.org/10.1074/jbc.M402379200
Plumley JA, Dannenberg JJ (2010) The importance of hydrogen bonding between the glutamine side chains to the formation of amyloid VQIVYK parallel beta-sheets: an ONIOM DFT/AM1 study. J Am Chem Soc 132:1758–1759. https://doi.org/10.1021/ja909690a
Li DW, Mohanty S, Irbäck A, Huo S (2008) Formation and growth of oligomers: a monte carlo study of an amyloid tau fragment. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1000238
Frenkel-Pinter M, Tal S, Scherzer-Attali R et al (2016) Naphthoquinone-tryptophan hybrid inhibits aggregation of the tau-derived peptide PHF6 and reduces neurotoxicity. J Alzheimers Dis 51:165–178. https://doi.org/10.3233/JAD-150927
Frenkel-Pinter M, Tal S, Scherzer-Attali R et al (2017) Cl-NQTrp alleviates tauopathy symptoms in a model organism through the inhibition of tau aggregation-engendered toxicity. Neurodegener Dis 17:73–82. https://doi.org/10.1159/000448518
KrishnaKumar VG, Paul A, Gazit E, Segal D (2018) Mechanistic insights into remodeled Tau-derived PHF6 peptide fibrils by Naphthoquinone-Tryptophan hybrids. Sci Rep 8:71. https://doi.org/10.1038/s41598-017-18443-2
KrishnaKumar VG, Gupta S (2017) Simplified method to obtain enhanced expression of tau protein from E. coli and one-step purification by direct boiling. Prep Biochem Biotechnol 47:530–538. https://doi.org/10.1080/10826068.2016.1275012
Frenkel-Pinter M, Richman M, Belostozky A et al (2016) Selective inhibition of aggregation and toxicity of a tau-derived peptide using its glycosylated analogues. Chemistry 22:5945–5952. https://doi.org/10.1002/chem.201504950
Mohamed T, Hoang T, Jelokhani-Niaraki M, Rao PPN (2013) Tau-derived-hexapeptide (306)VQIVYK(311) aggregation inhibitors: nitrocatechol moiety as a pharmacophore in drug design. ACS Chem Neurosci 4:1559–1570. https://doi.org/10.1021/cn400151a
Haj E, Losev Y, Guru KrishnaKumar V et al (2018) Integrating in vitro and in silico approaches to evaluate the “dual functionality” of palmatine chloride in inhibiting and disassembling Tau-derived VQIVYK peptide fibrils. Biochim Biophys Acta 1862:1565–1575. https://doi.org/10.1016/j.bbagen.2018.04.001
KrishnaKumar VG, Baweja L, Ralhan K, Gupta S (2018) Carbamylation promotes amyloidogenesis and induces structural changes in Tau-core hexapeptide fibrils. Biochim Biophys Acta Gen Subj 1862:2590–2604. https://doi.org/10.1016/j.bbagen.2018.07.030
Zheng J, Liu C, Sawaya MR et al (2011) Macrocyclic beta-sheet peptides that inhibit the aggregation of a tau-protein-derived hexapeptide. J Am Chem Soc 133:3144–3157. https://doi.org/10.1021/ja110545h
Calcul L, Zhang B, Jinwal UK et al (2012) Natural products as a rich source of tau-targeting drugs for Alzheimer’s disease. Future Med Chem 4:1751–1761. https://doi.org/10.4155/fmc.12.124
Hussain G, Rasul A, Anwar H et al (2018) Role of plant derived alkaloids and their mechanism in neurodegenerative disorders. Int J Biol Sci 14:341–357. https://doi.org/10.7150/ijbs.23247
Chua SW, Cornejo A, van Eersel J et al (2017) The polyphenol altenusin inhibits in vitro fibrillization of tau and reduces induced tau pathology in primary neurons. ACS Chem Neurosci 8:743–751. https://doi.org/10.1021/acschemneuro.6b00433
Dammers C, Yolcu D, Kukuk L et al (2016) Selection and characterization of tau binding d-enantiomeric peptides with potential for therapy of alzheimer disease. PLoS One 11:e0167432
Sievers SA, Karanicolas J, Chang HW et al (2011) Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475:96–100. https://doi.org/10.1038/nature10154
Scherzer-Attali R, Farfara D, Cooper I et al (2012) Naphthoquinone-tyrptophan reduces neurotoxic Abeta*56 levels and improves cognition in Alzheimer’s disease animal model. Neurobiol Dis 46:663–672. https://doi.org/10.1016/j.nbd.2012.03.005
Cecchelli R, Aday S, Sevin E et al (2014) A stable and reproducible human blood-brain barrier model derived from hematopoietic stem cells. PLoS One 9:e99733. https://doi.org/10.1371/journal.pone.0099733
Pickhardt M, Gazova Z, von Bergen M et al (2005) Anthraquinones inhibit tau aggregation and dissolve Alzheimer’s paired helical filaments in vitro and in cells. J Biol Chem 280:3628–3635. https://doi.org/10.1074/jbc.M410984200
Viswanathan GK, Mohapatra S, Paul A et al (2018) Inhibitory effect of naphthoquinone-tryptophan hybrid towards aggregation of PAP f39 semen amyloid. Molecules. https://doi.org/10.3390/molecules23123279
Sastry GM, Adzhigirey M, Day T et al (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/s10822-013-9644-8
Friesner RA, Murphy RB, Repasky MP et al (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177–6196. https://doi.org/10.1021/jm051256o
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J Med Chem 47:1739–1749. https://doi.org/10.1021/jm0306430
Halgren TA, Murphy RB, Friesner RA et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. enrichment factors in database screening. J Med Chem 47:1750–1759. https://doi.org/10.1021/jm030644s
Jacobson MP, Friesner RA, Xiang Z, Honig B (2002) On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol 320:597–608
Jacobson MP, Pincus DL, Rapp CS et al (2004) A hierarchical approach to all-atom protein loop prediction. Proteins 55:351–367. https://doi.org/10.1002/prot.10613
Farid R, Day T, Friesner RA, Pearlstein RA (2006) New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg Med Chem 14:3160–3173. https://doi.org/10.1016/j.bmc.2005.12.032
Sherman W, Beard HS, Farid R (2006) Use of an induced fit receptor structure in virtual screening. Chem Biol Drug Des 67:83–84. https://doi.org/10.1111/j.1747-0285.2005.00327.x
Sherman W, Day T, Jacobson MP et al (2006) Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49:534–553. https://doi.org/10.1021/jm050540c
Sawaya MR, Sambashivan S, Nelson R et al (2007) Atomic structures of amyloid cross-[beta] spines reveal varied steric zippers. Nature 447:453–457
Schuttelkopf AW, van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. https://doi.org/10.1107/S0907444904011679
Schmid N, Eichenberger AP, Choutko A et al (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J 40:843–856. https://doi.org/10.1007/s00249-011-0700-9
Lindahl E, Hess B, van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. Mol Model Annu 7:306–317. https://doi.org/10.1007/s008940100045
Gerben SR, Lemkul JA, Brown AM, Bevan DR (2014) Comparing atomistic molecular mechanics force fields for a difficult target: a case study on the Alzheimer’s amyloid beta-peptide. J Biomol Struct Dyn 32:1817–1832. https://doi.org/10.1080/07391102.2013.838518
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12%3c1463:AID-JCC4%3e3.0.CO;2-H
Goedert M, Spillantini MG, Jakes R et al (1989) Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3:519–526
Kolarova M, Garcia-Sierra F, Bartos A et al (2012) Structure and pathology of tau protein in Alzheimer disease. Int J Alzheimers Dis 2012:731526. https://doi.org/10.1155/2012/731526
Chatterjee S, Sang T-K, Lawless GM, Jackson GR (2009) Dissociation of tau toxicity and phosphorylation: role of GSK-3beta, MARK and Cdk5 in a Drosophila model. Hum Mol Genet 18:164–177. https://doi.org/10.1093/hmg/ddn326
Scherzer-Attali R, Pellarin R, Convertino M et al (2010) Complete phenotypic recovery of an Alzheimer’s disease model by a quinone-tryptophan hybrid aggregation inhibitor. PLoS One 5:e11101. https://doi.org/10.1371/journal.pone.0011101
Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11:36–42
Luo H, Gauthier M, Tan X et al (2018) Sodium transporters are involved in lithium influx in brain endothelial cells. Mol Pharm 15:2528–2538. https://doi.org/10.1021/acs.molpharmaceut.8b00018
Loureiro JA, Andrade S, Duarte A et al (2017) Resveratrol and grape extract-loaded solid lipid nanoparticles for the treatment of Alzheimer’s disease. Molecules. https://doi.org/10.3390/molecules22020277
Eigenmann DE, Durig C, Jahne EA et al (2016) In vitro blood-brain barrier permeability predictions for GABAA receptor modulating piperine analogs. Eur J Pharm Biopharm 103:118–126. https://doi.org/10.1016/j.ejpb.2016.03.029
Kuntz M, Candela P, Saint-Pol J et al (2015) Bexarotene promotes cholesterol efflux and restricts apical-to-basolateral transport of amyloid-beta peptides in an in vitro model of the human blood-brain barrier. J Alzheimers Dis 48:849–862. https://doi.org/10.3233/JAD-150469
Praca C, Rai A, Santos T et al (2018) A nanoformulation for the preferential accumulation in adult neurogenic niches. J Control Release 284:57–72. https://doi.org/10.1016/j.jconrel.2018.06.013
Cecchelli R, Dehouck B, Descamps L et al (1999) In vitro model for evaluating drug transport across the blood-brain barrier. Adv Drug Deliv Rev 36:165–178
Khurana R, Coleman C, Ionescu-Zanetti C et al (2005) Mechanism of thioflavin T binding to amyloid fibrils. J Struct Biol 151:229–238. https://doi.org/10.1016/j.jsb.2005.06.006
Groenning M (2010) Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils—current status. J Chem Biol 3:1–18. https://doi.org/10.1007/s12154-009-0027-5
Xue C, Lin TY, Chang D, Guo Z (2017) Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. R Soc Open Sci 4:160696. https://doi.org/10.1098/rsos.160696
Ross PD, Subramanian S (1981) Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry 20:3096–3102
Wang S-H, Liu F-F, Dong X-Y, Sun Y (2010) Thermodynamic analysis of the molecular interactions between amyloid beta-peptide 42 and (-)-epigallocatechin-3-gallate. J Phys Chem B 114:11576–11583. https://doi.org/10.1021/jp1001435
Berhanu WM, Masunov AE (2015) Atomistic mechanism of polyphenol amyloid aggregation inhibitors: molecular dynamics study of Curcumin, Exifone, and Myricetin interaction with the segment of tau peptide oligomer. J Biomol Struct Dyn 33:1399–1411. https://doi.org/10.1080/07391102.2014.951689
Zhao J-H, Liu H-L, Chuang C-K et al (2010) Molecular dynamics simulations to investigate the stability and aggregation behaviour of the amyloid-forming peptide VQIVYK from tau protein. Mol Simul 36:1013–1024. https://doi.org/10.1080/08927022.2010.499147
Velander P, Wu L, Henderson F et al (2017) Natural product-based amyloid inhibitors. Biochem Pharmacol 139:40–55. https://doi.org/10.1016/j.bcp.2017.04.004
Armstrong AH, Chen J, McKoy AF, Hecht MH (2011) Mutations that replace aromatic side chains promote aggregation of the Alzheimer’s Abeta peptide. Biochemistry 50:4058–4067. https://doi.org/10.1021/bi200268w
Sang T-K, Jackson GR (2005) Drosophila models of neurodegenerative disease. NeuroRx 2:438–446. https://doi.org/10.1602/neurorx.2.3.438
Moloney A, Sattelle DB, Lomas DA, Crowther DC (2010) Alzheimer’s disease: insights from Drosophila melanogaster models. Trends Biochem Sci 35:228–235. https://doi.org/10.1016/j.tibs.2009.11.004
Wittmann CW, Wszolek MF, Shulman JM et al (2001) Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science 293:711–714. https://doi.org/10.1126/science.1062382
Pandey UB, Nichols CD (2011) Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol Rev 63:411–436. https://doi.org/10.1124/pr.110.003293
Passarella D, Goedert M (2018) Beta-sheet assembly of Tau and neurodegeneration in Drosophila melanogaster. Neurobiol Aging 72:98–105. https://doi.org/10.1016/j.neurobiolaging.2018.07.022
Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415
Ephrussi B, Herold JL (1944) Studies of eye pigments of Drosophila. I. Methods of extraction and quantitative estimation of the pigment components. Genetics 29:148–175
Bancher C, Brunner C, Lassmann H et al (1989) Accumulation of abnormally phosphorylated tau precedes the formation of neurofibrillary tangles in Alzheimer’s disease. Brain Res 477:90–99
Ravid O, Elhaik Goldman S, Macheto D et al (2018) Blood-brain barrier cellular responses toward organophosphates: natural compensatory processes and exogenous interventions to rescue barrier properties. Front Cell Neurosci 12:359. https://doi.org/10.3389/fncel.2018.00359
Acknowledgements
This work was supported in part by the Alliance Family Foundation, and the Rosetrees Trust (to DS). This work was done in collaboration with the BLAVATNIK CENTER for Drug Discovery supported by the Blavatnik Family Foundation. GKV thanks TATA post-doctoral scholarship. Authors are grateful to the members of EG and DS research groups for fruitful discussions. We thank Donna Elyashiv Revivo for introduction and advice on fly work. Authors thank Dr. Vered Holdengreber for help with Electron Microscopy.
Author information
Authors and Affiliations
Contributions
GKV and DS conceived and designed the project. GKV and DSh conducted the lab experiments. LL contributed to the development of cell model and western blots. EA and RJ performed ITC and SPR assays. GKV, EP, and AR executed the high throughput screening assay. HE performed molecular docking. FG established the in vitro BBB model. CS and IC performed the BBB permeability assay. GKV, DSh, EG and DS prepared the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Viswanathan, G.K., Shwartz, D., Losev, Y. et al. Purpurin modulates Tau-derived VQIVYK fibrillization and ameliorates Alzheimer’s disease-like symptoms in animal model. Cell. Mol. Life Sci. 77, 2795–2813 (2020). https://doi.org/10.1007/s00018-019-03312-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03312-0