Abstract
The crosstalk between prokaryotic bacteria and eukaryotic gut epithelial cells has opened a new field for research. Quorum sensing system (QS) molecules employed by gut microbiota may play an essential role in host–microbial symbioses of the gut. Recent studies on the gut microbiome will unveil evolved mechanisms of the host to affect bacterial QS and shape bacterial composition. Bacterial autoinducers (AIs) could talk to the host’s gut by eliciting proinflammatory effects and modulating the activities of T lymphocyte, macrophage, dendritic cells, and neutrophils. In addition, the gut mucosa could interfere with bacterial AIs by degrading them or secreting AI mimics. Moreover, bacterial AIs and gut hormones epinephrine and noradrenaline may be interchangeable in the crosstalk between the microbiota and human gut. Therefore, inter-kingdom signaling between gut microbiota and host may provide a novel target in the management of gut microbiota-related conditions or diseases in the future.

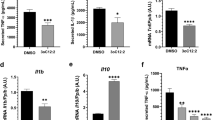
Modified from Ref. [35]

Similar content being viewed by others
References
Wu Y, Wu J, Chen T, Li Q, Peng W, Li H, Tang X, Fu X (2018) Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a toll-like receptor 4/p21-activated kinase 1 cascade. Dig Dis Sci 63(5):1210–1218
Pacheco AR, Sperandio V (2009) Inter-kingdom signaling: chemical language between bacteria and host. Curr Opin Microbiol 12(2):192–198
Shiner EK, Rumbaugh KP, Williams SC (2005) Inter-kingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol Rev 29(5):935–947
Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, Trinchieri G (2017) Microbes and cancer. Annu Rev Immunol 35:199–228
Hall AB, Tolonen AC, Xavier RJ (2017) Human genetic variation and the gut microbiome in disease. Nat Rev Genet 18(11):690–699
Maruvada P, Leone V, Kaplan LM, Chang EB (2017) The human microbiome and obesity: moving beyond associations. Cell Host Microbe 22(5):589–599
Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK et al (2015) Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528(7581):262–266
Scott TA, Quintaneiro LM, Norvaisas P, Lui PP, Wilson MP, Leung KY, Herrera-Dominguez L, Sudiwala S, Pessia A, Clayton PT et al (2017) Host–microbe co-metabolism dictates cancer drug efficacy in C. elegans. Cell 169(3):442 e418–456 e418
Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL et al (2013) Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14(2):207–215
Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, Li Q, Wu J, Fu X (2017) Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget 8(19):31802–31814
Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H et al (2017) The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8(1):845
Ost KS, Round JL (2017) A few good commensals: gut microbes use IFN-gamma to fight salmonella. Immunity 46(6):977–979
Belkaid Y, Hand TW (2014) Role of the microbiota in immunity and inflammation. Cell 157(1):121–141
Rook G, Bäckhed F, Levin BR, McFall-Ngai MJ, McLean AR (2017) Evolution, human–microbe interactions, and life history plasticity. Lancet 390(10093):521–530
Postler TS, Ghosh S (2017) Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab 26(1):110–130
Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM (2004) Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127(1):80–93
Hughes DT, Sperandio V (2008) Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol 6(2):111–120
Whiteley M, Diggle SP, Greenberg EP (2017) Progress in and promise of bacterial quorum sensing research. Nature 551(7680):313–320
Kaper JB, Sperandio V (2005) Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun 73(6):3197–3209
Moreno-Gamez S, Sorg RA, Domenech A, Kjos M, Weissing FJ, van Doorn GS, Veening JW (2017) Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun 8(1):854
Sintim HO, Smith JA, Wang J, Nakayama S, Yan L (2010) Paradigm shift in discovering next-generation anti-infective agents: targeting quorum sensing, c-di-GMP signaling and biofilm formation in bacteria with small molecules. Fut Med Chem 2(6):1005–1035
Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR (2005) Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 3(5):383–396
Xavier KB, Bassler BL (2005) Interference with AI-2-mediated bacterial cell–cell communication. Nature 437(7059):750–753
Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB (2015) Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep 10(11):1861–1871
Galloway WR, Hodgkinson JT, Bowden SD, Welch M, Spring DR (2011) Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev 111(1):28–67
Ismail AS, Valastyan JS, Bassler BL (2016) A host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe 19(4):470–480
Elmanfi S, Ma X, Sintim HO, Kononen E, Syrjanen S, Gursoy UK (2018) Quorum-sensing molecule dihydroxy-2,3-pentanedione and its analogs as regulators of epithelial integrity. J Periodontal Res 53(3):414–421
Zargar A, Quan DN, Carter KK, Guo M, Sintim HO, Payne GF, Bentley WE (2015) Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. mBio 6(2):e00025
Smith RS, Harris SG, Phipps R, Iglewski B (2002) The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol 184(4):1132–1139
Scheres N, Lamont RJ, Crielaard W, Krom BP (2015) LuxS signaling in Porphyromonas gingivalis-host interactions. Anaerobe 35(Pt A):3–9
Cui ZQ, Wu ZM, Fu YX, Xu DX, Guo X, Shen HQ, Wei XB, Yi PF, Fu BD (2016) Autoinducer-2 of quorum sensing is involved in cell damage caused by avian pathogenic Escherichia coli. Microb Pathog 99:247–252
Vidal JE, Howery KE, Ludewick HP, Nava P, Klugman KP (2013) Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun 81(4):1341–1353
Trier JS (2002) Mucosal flora in inflammatory bowel disease: intraepithelial bacteria or endocrine epithelial cell secretory granules? Gastroenterology 123(3):955 (author reply 956)
Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M et al (2014) Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA 111(51):18321–18326
Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y (2016) Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer 139(6):1318–1326
Clark JA, Coopersmith CM (2007) Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock 28(4):384–393
Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI (2011) Human nutrition, the gut microbiome and the immune system. Nature 474(7351):327–336
Tateda K, Ishii Y, Horikawa M, Matsumoto T, Miyairi S, Pechere JC, Standiford TJ, Ishiguro M, Yamaguchi K (2003) The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect Immun 71(10):5785–5793
Shiner EK, Terentyev D, Bryan A, Sennoune S, Martinez-Zaguilan R, Li G, Gyorke S, Williams SC, Rumbaugh KP (2006) Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell Microbiol 8(10):1601–1610
Telford G, Wheeler D, Williams P, Tomkins PT, Appleby P, Sewell H, Stewart GS, Bycroft BW, Pritchard DI (1998) The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect Immun 66(1):36–42
Ritchie AJ, Yam AO, Tanabe KM, Rice SA, Cooley MA (2003) Modification of in vivo and in vitro T- and B-cell-mediated immune responses by the Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)-l-homoserine lactone. Infect Immun 71(8):4421–4431
Chhabra SR, Harty C, Hooi DS, Daykin M, Williams P, Telford G, Pritchard DI, Bycroft BW (2003) Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J Med Chem 46(1):97–104
Skindersoe ME, Zeuthen LH, Brix S, Fink LN, Lazenby J, Whittall C, Williams P, Diggle SP, Froekiaer H, Cooley M et al (2009) Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol Med Microbiol 55(3):335–345
Ritchie AJ, Whittall C, Lazenby JJ, Chhabra SR, Pritchard DI, Cooley MA (2007) The immunomodulatory Pseudomonas aeruginosa signalling molecule N-(3-oxododecanoyl)-l-homoserine lactone enters mammalian cells in an unregulated fashion. Immunol Cell Biol 85(8):596–602
Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Grauer DC, Lehmann M, Meijler MM, Janda KD, Ulevitch RJ (2008) Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science 321(5886):259–263
Wynendaele E, Verbeke F, D’Hondt M, Hendrix A, Van De Wiele C, Burvenich C, Peremans K, De Wever O, Bracke M, De Spiegeleer B (2015) Crosstalk between the microbiome and cancer cells by quorum sensing peptides. Peptides 64:40–48
Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP (2008) Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol 190(13):4408–4415
Hooper LV, Littman DR, Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336(6086):1268–1273
Yeo S, Park H, Ji Y, Park S, Yang J, Lee J, Mathara JM, Shin H, Holzapfel W (2015) Influence of gastrointestinal stress on autoinducer-2 activity of two Lactobacillus species. FEMS Microbiol Ecol 91(7):fiv065
Chun CK, Ozer EA, Welsh MJ, Zabner J, Greenberg EP (2004) Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc Natl Acad Sci USA 101(10):3587–3590
Teiber JF, Horke S, Haines DC, Chowdhary PK, Xiao J, Kramer GL, Haley RW, Draganov DI (2008) Dominant role of paraoxonases in inactivation of the Pseudomonas aeruginosa quorum-sensing signal N-(3-oxododecanoyl)-l-homoserine lactone. Infect Immun 76(6):2512–2519
Mathesius U, Mulders S, Gao M, Teplitski M, Caetano-Anolles G, Rolfe BG, Bauer WD (2003) Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc Natl Acad Sci USA 100(3):1444–1449
Eisenhofer G, Aneman A, Friberg P, Hooper D, Fandriks L, Lonroth H, Hunyady B, Mezey E (1997) Substantial production of dopamine in the human gastrointestinal tract. J Clin Endocrinol Metab 82(11):3864–3871
Lyte M, Frank CD, Green BT (1996) Production of an autoinducer of growth by norepinephrine cultured Escherichia coli O157:H7. FEMS Microbiol Lett 139(2–3):155–159
Williams SC, Patterson EK, Carty NL, Griswold JA, Hamood AN, Rumbaugh KP (2004) Pseudomonas aeruginosa autoinducer enters and functions in mammalian cells. J Bacteriol 186(8):2281–2287
Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB (2003) Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA 100(15):8951–8956
Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, Ciancio M, Zaborin A, Petrof EO, Turner JR et al (2007) Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog 3(3):e35
Chen T, Li Q, Zhang X, Long R, Wu Y, Wu J, Fu X (2018) TOX expression decreases with progression of colorectal cancers and is associated with CD4 T-cell density and Fusobacterium nucleatum infection. Hum Pathol 79:93–101
Chen T, Li Q, Wu J, Wu Y, Peng W, Li H, Wang J, Tang X, Peng Y, Fu X (2018) Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol Immunother CII 67(10):1635–1646
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interests for this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Q., Ren, Y. & Fu, X. Inter-kingdom signaling between gut microbiota and their host. Cell. Mol. Life Sci. 76, 2383–2389 (2019). https://doi.org/10.1007/s00018-019-03076-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03076-7