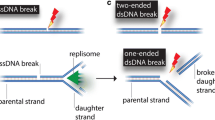
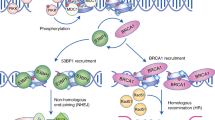
Abstract
Double-strand breaks (DSBs) are the most detrimental form of DNA damage. Failure to repair these cytotoxic lesions can result in genome rearrangements conducive to the development of many diseases, including cancer. The DNA damage response (DDR) ensures the rapid detection and repair of DSBs in order to maintain genome integrity. Central to the DDR are the DNA damage checkpoints. When activated by DNA damage, these sophisticated surveillance mechanisms induce transient cell cycle arrests, allowing sufficient time for DNA repair. Since the term “checkpoint” was coined over 20 years ago, our understanding of the molecular mechanisms governing the DNA damage checkpoint has advanced significantly. These pathways are highly conserved from yeast to humans. Thus, significant findings in yeast may be extrapolated to vertebrates, greatly facilitating the molecular dissection of these complex regulatory networks. This review focuses on the cellular response to DSBs in Saccharomyces cerevisiae, providing a comprehensive overview of how these signalling pathways function to orchestrate the cellular response to DNA damage and preserve genome stability in eukaryotic cells.




Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Abbreviations
- ATM:
-
Ataxia telangiectasia mutated
- ATR:
-
ATM and Rad3-related
- ATRIP:
-
ATR interacting protein
- BRCT:
-
BRCA1 carboxyl terminal
- CAD:
-
Chk1 activation domain
- CDK:
-
Cyclin-dependent kinase
- DDK:
-
Dbf4-dependent kinase
- DDR:
-
DNA damage response
- DNA-PKcs:
-
DNA-dependent protein kinase catalytic subunit
- DSB:
-
Double-strand break
- GCRs:
-
Gross chromosomal rearrangements
- G1:
-
Gap phase 1
- G2:
-
Gap phase 2
- HR:
-
Homologous recombination
- IR:
-
Ionising radiation
- M:
-
Mitosis
- MEN:
-
Mitotic exit network
- MMS:
-
Methyl methanesulfonate
- MRX/MRN:
-
Mre11-Rad50-Xrs2/MRE11-RAD50-NBS1
- NHEJ:
-
Non-homologous end joining
- PIKK:
-
Phosphoinositide 3-kinase related kinase
- PTM:
-
Post-translational modification
- RNR:
-
Ribonucleotide reductase
- RPA:
-
Replication protein A
- SCD:
-
SQ/TQ cluster domain
- UV:
-
Ultraviolet
References
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362(6422):709–715
Elledge SJ, Ciccia A (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40(2):179–204. doi:10.1016/j.molcel.2010.09.019
Hartwell LH, Weinert TA (1989) Checkpoints: controls that ensure the order of cell cycle events. Science 246(4930):629–634
Weinert TA, Hartwell LH (1988) The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241(4863):317–322
Lowndes NF, Murguia JR (2000) Sensing and responding to DNA damage. Curr Opin Genet Dev 10(1):17–25
Zhou BB, Elledge SJ (2000) The DNA damage response: putting checkpoints in perspective. Nature 408(6811):433–439
Bartek J, Lukas J (2007) DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 19(2):238–245
Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28(5):739–745
Clemenson C, Marsolier-Kergoat MC (2009) DNA damage checkpoint inactivation: adaptation and recovery. DNA Repair (Amst) 8(9):1101–1109
Weinberg RA, Hanahan D (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674. doi:10.1016/j.cell.2011.02.013
Bartek J, Jackson SP (2009) The DNA-damage response in human biology and disease. Nature 461(7267):1071–1078. doi:10.1038/nature08467
Hartwell LH, Unger MW (1977) Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol 75(2 Pt 1):422–435
Weinert T, Hartwell L (1989) Control of G2 delay by the rad9 gene of Saccharomyces cerevisiae. J Cell Sci Suppl 12:145–148
Hartwell LH, Mortimer RK, Culotti J, Culotti M (1973) Genetic control of the cell division cycle in yeast: V. genetic analysis of cdc mutants. Genetics 74(2):267–286
Lisby M, Rothstein R (2009) Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 8(9):1068–1076
Morgan DO (2007) The cell cycle: principles of control. Primers in biology. New Science, London
Futcher B (1996) Cyclins and the wiring of the yeast cell cycle. Yeast 12(16):1635–1646
Bahler J (2005) Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet 39:69–94
Forsburg SL, Nurse P (1991) Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annu Rev Cell Biol 7:227–256. doi:10.1146/annurev.cb.07.110191.001303
Lim HH, Goh PY, Surana U (1996) Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol 16(11):6385–6397
Yamamoto A, Guacci V, Koshland D (1996) Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J Cell Biol 133(1):99–110
Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425(6960):859–864
Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325(5948):1682–1686
Enserink JM, Kolodner RD (2010) An overview of Cdk1-controlled targets and processes. Cell Div 5:11
Hartwell LH, Culotti J, Reid B (1970) Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci USA 66(2):352–359
Hartwell LH, Culotti J, Pringle JR, Reid BJ (1974) Genetic control of the cell division cycle in yeast. Science 183(4120):46–51
Fitz Gerald JN, Benjamin JM, Kron SJ (2002) Robust G1 checkpoint arrest in budding yeast: dependence on DNA damage signaling and repair. J Cell Sci 115(Pt 8):1749–1757
Siede W, Friedberg AS, Dianova I, Friedberg EC (1994) Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics 138(2):271–281
Siede W, Friedberg AS, Friedberg EC (1993) RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 90(17):7985–7989
Siede W, Allen JB, Elledge SJ, Friedberg EC (1996) The Saccharomyces cerevisiae MEC1 gene, which encodes a homolog of the human ATM gene product, is required for G1 arrest following radiation treatment. J Bacteriol 178(19):5841–5843
Longhese MP, Clerici M, Lucchini G (2003) The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat Res 532(1–2):41–58
Paulovich AG, Hartwell LH (1995) A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82(5):841–847
Lindahl T (2000) Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res 462(2-3):129–135 S1383574200000247 [pii]
Harrison JC, Haber JE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40:209–235
Lovejoy CA, Cortez D (2009) Common mechanisms of PIKK regulation. DNA Repair (Amst) 8(9):1004–1008
Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23(10):394–398
Suzuki K, Kodama S, Watanabe M (1999) Recruitment of ATM protein to double strand DNA irradiated with ionizing radiation. J Biol Chem 274(36):25571–25575
Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300(5625):1542–1548
Dart DA, Adams KE, Akerman I, Lakin ND (2004) Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S-phase. J Biol Chem 279(16):16433–16440. doi:10.1074/jbc.M314212200
Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O (2006) ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med 203(2):297–303. doi:10.1084/jem.20051923
Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8(1):37–45
Adams KE, Medhurst AL, Dart D, Lakin ND (2006) Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 25(28):3894–3904. doi:10.1038/sj.onc.1209426
Stracker TH, Usui T, Petrini JH (2009) Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 8(9):1047–1054
Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ (1999) Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286(5442):1166–1171
Branzei D, Foiani M (2008) Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9(4):297–308
Bensimon A, Aebersold R, Shiloh Y (2011) Beyond ATM: the protein kinase landscape of the DNA damage response. FEBS Lett 585(11):1625–1639. doi:10.1016/j.febslet.2011.05.013
Polo SE, Jackson SP (2011) Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25(5):409–433. doi:10.1101/gad.2021311
Bennett CB, Lewis AL, Baldwin KK, Resnick MA (1993) Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci USA 90(12):5613–5617
Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63(2):349–404
Wu D, Topper LM, Wilson TE (2008) Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178(3):1237–1249
Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97(5):621–633
Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118(6):699–713
Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP (2008) The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9(8):810–818
Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE (2007) Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol 14(7):639–646
Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29(19):3370–3380. doi:10.1038/emboj.2010.219
Walker JR, Corpina RA, Goldberg J (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412(6847):607–614
Gravel S, Larrivee M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280(5364):741–744
Gottlieb TM, Jackson SP (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72(1):131–142
Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O (2006) Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 22(4):511–519
Weterings E, Verkaik NS, Bruggenwirth HT, Hoeijmakers JH, van Gent DC (2003) The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res 31(24):7238–7246
Hiom K (2010) Coping with DNA double strand breaks. DNA Repair (Amst) 9(12):1256–1263. doi:10.1016/j.dnarep.2010.09.018
Smith GC, Divecha N, Lakin ND, Jackson SP (1999) DNA-dependent protein kinase and related proteins. Biochem Soc Symp 64:91–104
Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8(5):1105–1115
Wiltzius JJ, Hohl M, Fleming JC, Petrini JH (2005) The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol 12(5):403–407
Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA (2002) The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418(6897):562–566
Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, Strasser K, Hopfner KP (2011) The Mre11: Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145(1):54–66. doi:10.1016/j.cell.2011.02.038
Frank-Vaillant M, Marcand S (2002) Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell 10(5):1189–1199
Barlow JH, Lisby M, Rothstein R (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30(1):73–85
Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431(7011):1011–1017
Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23(24):4868–4875
Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455(7213):689–692
Kadyk LC, Hartwell LH (1993) Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics 133(3):469–487
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. doi:10.1146/annurev.biochem.052308.093131
Heyer WD, Ehmsen KT, Liu J (2010) Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44:113–139. doi:10.1146/annurev-genet-051710-150955
Rupnik A, Lowndes NF, Grenon M (2010) MRN and the race to the break. Chromosoma 119(2):115–135. doi:10.1007/s00412-009-0242-4
Nakada D, Matsumoto K, Sugimoto K (2003) ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 17(16):1957–1962
Usui T, Ogawa H, Petrini JH (2001) A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell 7(6):1255–1266
D’Amours D, Jackson SP (2001) The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev 15(17):2238–2249
Grenon M, Gilbert C, Lowndes NF (2001) Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat Cell Biol 3(9):844–847
Mantiero D, Clerici M, Lucchini G, Longhese MP (2007) Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep 8(4):380–387
Vialard JE, Gilbert CS, Green CM, Lowndes NF (1998) The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J 17(19):5679–5688
Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P (1995) TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82(5):831–840
Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271(5247):357–360
Fukunaga K, Kwon Y, Sung P, Sugimoto K (2011) Activation of protein kinase Tel1 through recognition of protein-bound DNA ends. Mol Cell Biol 31(10):1959–1971. doi:10.1128/MCB.05157-11
Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP (2004) The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol 24(10):4151–4165
Clerici M, Mantiero D, Lucchini G, Longhese MP (2006) The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep 7(2):212–218
Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421(6922):499–506. doi:10.1038/nature01368
Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434(7033):605–611
You Z, Chahwan C, Bailis J, Hunter T, Russell P (2005) ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25(13):5363–5379
Lee JH, Paull TT (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308(5721):551–554. doi:10.1126/science.1108297
Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V (2008) Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J 27(14):1953–1962. doi:10.1038/emboj.2008.128
Shiotani B, Zou L (2009) Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33(5):547–558. doi:10.1016/j.molcel.2009.01.024
Sun Y, Xu Y, Roy K, Price BD (2007) DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol 27(24):8502–8509. doi:10.1128/MCB.01382-07
Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 102(37):13182–13187. doi:10.1073/pnas.0504211102
Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276(45):42462–42467
Downs JA, Lowndes NF, Jackson SP (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408(6815):1001–1004
Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA (2004) ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 64(7):2390–2396
Ward IM, Chen J (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276(51):47759–47762
Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M (2004) Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14(19):1703–1711
Downs JA, Nussenzweig MC, Nussenzweig A (2007) Chromatin dynamics and the preservation of genetic information. Nature 447(7147):951–958
Costelloe T, Fitzgerald J, Murphy NJ, Flaus A, Lowndes NF (2006) Chromatin modulation and the DNA damage response. Exp Cell Res 312(14):2677–2686
Rossetto D, Truman AW, Kron SJ, Cote J (2010) Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res 16(18):4543–4552. doi:10.1158/1078-0432.CCR-10-0513
Xu Y, Price BD (2011) Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle 10(2):261–267
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273(10):5858–5868
Rogakou EP, Boon C, Redon C, Bonner WM (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146(5):905–916
Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM (2003) Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep 4(7):1–7
Toh GW, O’Shaughnessy AM, Jimeno S, Dobbie IM, Grenon M, Maffini S, O’Rorke A, Lowndes NF (2006) Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 5:693–703
Hammet A, Magill C, Heierhorst J, Jackson SP (2007) Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep 8(9):851–857
Javaheri A, Wysocki R, Jobin-Robitaille O, Altaf M, Cote J, Kron SJ (2006) Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci USA 103(37):13771–13776
Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW (2003) Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114(3):359–370
Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A (2003) H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114(3):371–383
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A (2002) Genomic instability in mice lacking histone H2AX. Science 296(5569):922–927
Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R (2004) Control of sister chromatid recombination by histone H2AX. Mol Cell 16(6):1017–1025
Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, Carpenter PB, Bonner WM, Chen J, Nussenzweig A (2002) DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol 4(12):993–997
van Attikum H, Gasser SM (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19(5):207–217
Huertas D, Sendra R, Munoz P (2009) Chromatin dynamics coupled to DNA repair. Epigenetics 4(1):31–42
Soutoglou E, Misteli T (2008) Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320(5882):1507–1510
Bonilla CY, Melo JA, Toczyski DP (2008) Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell 30(3):267–276
Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5(7):675–679
Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J (2006) MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21(2):187–200
Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123(7):1213–1226
Al-Hakim A, Escribano-Diaz C, Landry MC, O’Donnell L, Panier S, Szilard RK, Durocher D (2010) The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 9(12):1229–1240. doi:10.1016/j.dnarep.2010.09.011
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131(5):887–900
Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131(5):901–914
Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318(5856):1637–1640
Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136(3):420–434
Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136(3):435–446
Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276(18):14537–14540
Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N (2010) HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12(1):80–86; sup pp 81–12. doi:10.1038/ncb2008
Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S (2009) RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res 37(7):2176–2193. doi:10.1093/nar/gkp082
Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455(7214):770–774
Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134(6):981–994
Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108(2):183–193
Deng C, Brown JA, You D, Brown JM (2005) Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170(2):591–600
Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28(4):638–651
Clerici M, Mantiero D, Lucchini G, Longhese MP (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280(46):38631–38638
Dynan WS, Yoo S (1998) Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res 26(7):1551–1559
Mimitou EP, Symington LS (2011) DNA end resection-Unraveling the tail. DNA Repair (Amst). doi:10.1016/j.dnarep.2010.12.004
Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22(20):2767–2772
Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT (2010) Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol 17(12):1478–1485. doi:10.1038/nsmb.1957
Mimitou EP, Symington LS (2010) Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29(19):3358–3369. doi:10.1038/emboj.2010.193
Engels K, Giannattasio M, Muzi-Falconi M, Lopes M, Ferrari S (2011) 14-3-3 Proteins regulate exonuclease 1-dependent processing of stalled replication forks. PLoS Genet 7(4):e1001367. doi:10.1371/journal.pgen.1001367
Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P (2010) Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467(7311):108–111. doi:10.1038/nature09318
Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC (2010) DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467(7311):112–116. doi:10.1038/nature09355
Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25(4):350–362. doi:10.1101/gad.2003811
Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC (2008) Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA 105(44):16906–16911. doi:10.1073/pnas.0809380105
Niu H, Raynard S, Sung P (2009) Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev 23(13):1481–1486
Longhese MP, Bonetti D, Manfrini N, Clerici M (2010) Mechanisms and regulation of DNA end resection. EMBO J 29(17):2864–2874. doi:10.1038/emboj.2010.165
Zierhut C, Diffley JF (2008) Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J 27(13):1875–1885. doi:10.1038/emboj.2008.111
Chen L, Nievera CJ, Lee AY, Wu X (2008) Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem 283(12):7713–7720
Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450(7169):509–514
Yu X, Chen J (2004) DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol 24(21):9478–9486
Yu X, Fu S, Lai M, Baer R, Chen J (2006) BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev 20(13):1721–1726
Yun MH, Hiom K (2009) CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459(7245):460–463
Huertas P, Jackson SP (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284(14):9558–9565
Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G (2011) Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol 18(9):1015–1019. doi:10.1038/nsmb.2105
Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94(3):399–409
Lydall D, Weinert T (1995) Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270(5241):1488–1491
Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M (2008) Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 27(10):1502–1512
Postow L (2011) Destroying the ring: Freeing DNA from Ku with ubiquitin. FEBS Lett 585(18):2876–2882. doi:10.1016/j.febslet.2011.05.046
Couto CA, Wang HY, Green JC, Kiely R, Siddaway R, Borer C, Pears CJ, Lakin ND (2011) PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks. J Cell Biol 194(3):367–375. doi:10.1083/jcb.201012132
Paciotti V, Clerici M, Lucchini G, Longhese MP (2000) The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev 14(16):2046–2059
Majka J, Niedziela-Majka A, Burgers PM (2006) The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24(6):891–901
Cortez D, Guntuku S, Qin J, Elledge SJ (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294(5547):1713–1716
Rouse J, Jackson SP (2002) Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell 9(4):857–869
Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K (2001) Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294(5543):867–870
Melo JA, Cohen J, Toczyski DP (2001) Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev 15(21):2809–2821
Itakura E, Takai KK, Umeda K, Kimura M, Ohsumi M, Tamai K, Matsuura A (2004) Amino-terminal domain of ATRIP contributes to intranuclear relocation of the ATR-ATRIP complex following DNA damage. FEBS Lett 577(1–2):289–293
Wu X, Shell SM, Zou Y (2005) Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene 24(29):4728–4735
Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH (2009) Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex–implications for clamp loading and regulation. Mol Cell 34(6):735–745
Sohn SY, Cho Y (2009) Crystal structure of the human rad9-hus1-rad1 clamp. J Mol Biol 390(3):490–502
Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF (2000) A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol 10(1):39–42
Majka J, Burgers PM (2003) Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA 100(5):2249–2254
Majka J, Binz SK, Wold MS, Burgers PM (2006) Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem 281(38):27855–27861
Ellison V, Stillman B (2003) Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol 1(2):E33
Cimprich KA (2007) Probing ATR activation with model DNA templates. Cell Cycle 6(19):2348–2354
Navadgi-Patil VM, Burgers PM (2009) The unstructured C-terminal tail of the 9–1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell 36(5):743–753
Paciotti V, Lucchini G, Plevani P, Longhese MP (1998) Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J 17(14):4199–4209
de la Torre-Ruiz MA, Green CM, Lowndes NF (1998) RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J 17(9):2687–2698
Brush GS, Kelly TJ (2000) Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res 28(19):3725–3732
Navadgi-Patil VM, Burgers PM (2011) Cell-cycle-specific activators of the Mec1/ATR checkpoint kinase. Biochem Soc Trans 39(2):600–605. doi:10.1042/BST0390600
Navadgi-Patil VM, Burgers PM (2009) A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9–1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst) 8(9):996–1003
Lee J, Kumagai A, Dunphy WG (2007) The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 282(38):28036–28044
Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM (2007) The Rad9-Hus1-Rad1 (9–1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21(12):1472–1477
Takeishi Y, Ohashi E, Ogawa K, Masai H, Obuse C, Tsurimoto T (2010) Casein kinase 2-dependent phosphorylation of human Rad9 mediates the interaction between human Rad9-Hus1-Rad1 complex and TopBP1. Genes Cells 15(7):761–771. doi:10.1111/j.1365-2443.2010.01418.x
Rappas M, Oliver AW, Pearl LH (2011) Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res 39(1):313–324. doi:10.1093/nar/gkq743
Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124(5):943–955
Mordes DA, Glick GG, Zhao R, Cortez D (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22(11):1478–1489
Mordes DA, Nam EA, Cortez D (2008) Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci USA 105(48):18730–18734
Navadgi-Patil VM, Burgers PM (2008) Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283(51):35853–35859
Pfander B, Diffley JF (2011) Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J. doi:10.1038/emboj.2011.345
Schwartz MF, Duong JK, Sun Z, Morrow JS, Pradhan D, Stern DF (2002) Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell 9(5):1055–1065
Emili A (1998) MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell 2(2):183–189
Gilbert CS, van den Bosch M, Green CM, Vialard JE, Grenon M, Erdjument-Bromage H, Tempst P, Lowndes NF (2003) The budding yeast Rad9 checkpoint complex: chaperone proteins are required for its function. EMBO Rep 4(10):953–958
Gilbert CS, Green CM, Lowndes NF (2001) Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell 8(1):129–136
Granata M, Lazzaro F, Novarina D, Panigada D, Puddu F, Abreu CM, Kumar R, Grenon M, Lowndes NF, Plevani P, Muzi-Falconi M (2010) Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet 6(8):e1001047
Grenon M, Costelloe T, Jimeno S, O’Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF (2007) Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24(2):105–119
Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432(7015):406–411
Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M (2008) Phosphorylation of the budding yeast 9–1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 28(15):4782–4793
van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109(6):745–756
Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J (2002) Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem 277(34):30421–30424
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389(6648):251–260
Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ (2005) Role of dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25(19):8430–8443
Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280(11):9879–9886
Lancelot N, Charier G, Couprie J, Duband-Goulet I, Alpha-Bazin B, Quemeneur E, Ma E, Marsolier-Kergoat MC, Ropars V, Charbonnier JB, Miron S, Craescu CT, Callebaut I, Gilquin B, Zinn-Justin S (2007) The checkpoint Saccharomyces cerevisiae Rad9 protein contains a tandem tudor domain that recognizes DNA. Nucleic Acids Res 35(17):5898–5912
Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D (2005) Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 15(15):1364–1375
Du LL, Nakamura TM, Russell P (2006) Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20(12):1583–1596
Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119(5):603–614
Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y (2002) Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 12(13):1086–1099
Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D (2002) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9(6):1201–1213
Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G (2006) Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell 127(7):1361–1373
Greeson NT, Sengupta R, Arida AR, Jenuwein T, Sanders SL (2008) Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J Biol Chem 283(48):33168–33174
Kilkenny ML, Dore AS, Roe SM, Nestoras K, Ho JC, Watts FZ, Pearl LH (2008) Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev 22(15):2034–2047
Nakamura TM, Du LL, Redon C, Russell P (2004) Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24(14):6215–6230
Nakamura TM, Moser BA, Du LL, Russell P (2005) Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol Cell Biol 25(24):10721–10730
Furuya K, Poitelea M, Guo L, Caspari T, Carr AM (2004) Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev 18(10):1154–1164
Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M (1997) Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev 11(24):3387–3400
Mochida S, Esashi F, Aono N, Tamai K, O’Connell MJ, Yanagida M (2004) Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J 23(2):418–428
Wang H, Elledge SJ (2002) Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics 160(4):1295–1304
Germann SM, Oestergaard VH, Haas C, Salis P, Motegi A, Lisby M (2011) Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair (Amst) 10(2):210–224. doi:10.1016/j.dnarep.2010.11.001
Lee J, Dunphy WG (2010) Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell 21(6):926–935. doi:10.1091/mbc.E09-11-0958
Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7(7):1389–1396
Smolka MB, Albuquerque CP, Chen SH, Schmidt KH, Wei XX, Kolodner RD, Zhou H (2005) Dynamic changes in protein–protein interaction and protein phosphorylation probed with amine-reactive isotope tag. Mol Cell Proteomics 4(9):1358–1369
Ward IM, Minn K, van Deursen J, Chen J (2003) p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23(7):2556–2563
Wang B, Matsuoka S, Carpenter PB, Elledge SJ (2002) 53BP1, a mediator of the DNA damage checkpoint. Science 298(5597):1435–1438
DiTullio RA Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD (2002) 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 4(12):998–1002
Wilson KA, Stern DF (2008) NFBD1/MDC1, 53BP1 and BRCA1 have both redundant and unique roles in the ATM pathway. Cell Cycle 7(22):3584–3594
Lee JH, Goodarzi AA, Jeggo PA, Paull TT (2010) 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J 29(3):574–585
Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470(7332):124–128. doi:10.1038/nature09658
Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421(6926):961–966
Xu X, Stern DF (2003) NFBD1/MDC1 regulates ionizing radiation-induced focus formation by DNA checkpoint signaling and repair factors. FASEB J 17(13):1842–1848. doi:10.1096/fj.03-0310com
Cotta-Ramusino C, McDonald ER 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ (2011) A DNA damage response screen identifies RHINO, a 9–1-1 and TopBP1 interacting protein required for ATR signaling. Science 332(6035):1313–1317. doi:10.1126/science.1203430
Yamane K, Wu X, Chen J (2002) A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol 22(2):555–566
Cescutti R, Negrini S, Kohzaki M, Halazonetis TD (2010) TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint. EMBO J 29(21):3723–3732. doi:10.1038/emboj.2010.238
Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP (2008) A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 105(31):10762–10767
Jowsey P, Morrice NA, Hastie CJ, McLauchlan H, Toth R, Rouse J (2007) Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair (Amst) 6(10):1536–1544
Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5(2):214–226
Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127(3):635–648
Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ (2007) Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci USA 104(50):19855–19860
Molina H, Horn DM, Tang N, Mathivanan S, Pandey A (2007) Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA 104(7):2199–2204
Tao WA, Wollscheid B, O’Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R (2005) Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods 2(8):591–598
Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Bohmer FD, Gerke V, Schmidt-Arras DE, Berdel WE, Muller-Tidow C, Mann M, Serve H (2009) Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell 36(2):326–339. doi:10.1016/j.molcel.2009.09.019
Chen Y, Farmer AA, Chen CF, Jones DC, Chen PL, Lee WH (1996) BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner. Cancer Res 56(14):3168–3172
Ruffner H, Verma IM (1997) BRCA1 is a cell cycle-regulated nuclear phosphoprotein. Proc Natl Acad Sci USA 94(14):7138–7143
Ruffner H, Jiang W, Craig AG, Hunter T, Verma IM (1999) BRCA1 is phosphorylated at serine 1497 in vivo at a cyclin-dependent kinase 2 phosphorylation site. Mol Cell Biol 19(7):4843–4854
Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE (2008) Identification of CDK2 substrates in human cell lysates. Genome Biol 9(10):R149
Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR 3rd (2008) Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res 7(3):1346–1351
Sun Z, Hsiao J, Fay DS, Stern DF (1998) Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281(5374):272–274
Durocher D, Henckel J, Fersht AR, Jackson SP (1999) The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4(3):387–394
Schwartz MF, Lee S, Duong JK, Eminaga S, Stern DF (2003) FHA domain-mediated DNA checkpoint regulation of Rad53. Cell Cycle 2(4):384–396
Smith J, Tho LM, Xu N, Gillespie DA (2010) The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 108:73–112. doi:10.1016/B978-0-12-380888-2.00003-0
Usui T, Foster SS, Petrini JH (2009) Maintenance of the DNA-damage checkpoint requires DNA-damage-induced mediator protein oligomerization. Mol Cell 33(2):147–159
Blankley RT, Lydall D (2004) A domain of Rad9 specifically required for activation of Chk1 in budding yeast. J Cell Sci 117(Pt 4):601–608
Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403(6770):623–627
Chen Y, Caldwell JM, Pereira E, Baker RW, Sanchez Y (2009) ATRMec1 phosphorylation-independent activation of Chk1 in vivo. J Biol Chem 284(1):182–190
Tapia-Alveal C, Calonge TM, O’Connell MJ (2009) Regulation of chk1. Cell Div 4:8
Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3(11):958–965
Foss EJ (2001) Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157(2):567–577
Osborn AJ, Elledge SJ (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17(14):1755–1767
Smolka MB, Chen SH, Maddox PS, Enserink JM, Albuquerque CP, Wei XX, Desai A, Kolodner RD, Zhou H (2006) An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J Cell Biol 175(5):743–753
Chen SH, Zhou H (2009) Reconstitution of Rad53 activation by Mec1 through adaptor protein Mrc1. J Biol Chem 284(28):18593–18604. doi:10.1074/jbc.M109.018242
Katsuragi Y, Sagata N (2004) Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol Biol Cell 15(4):1680–1689. doi:10.1091/mbc.E03-12-0874
Oe T, Nakajo N, Katsuragi Y, Okazaki K, Sagata N (2001) Cytoplasmic occurrence of the Chk1/Cdc25 pathway and regulation of Chk1 in Xenopus oocytes. Dev Biol 229(1):250–261. doi:10.1006/dbio.2000.9968
Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, Tempczyk-Russell A, Nguyen B, Myers P, Lundgren K, Kan CC, O’Connor PM, O’Connor PM (2000) The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell 100(6):681–692
Walker M, Black EJ, Oehler V, Gillespie DA, Scott MT (2009) Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene 28(24):2314–2323. doi:10.1038/onc.2009.102
Smits VA, Reaper PM, Jackson SP (2006) Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol 16(2):150–159. doi:10.1016/j.cub.2005.11.066
Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M (2008) Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132(2):221–232. doi:10.1016/j.cell.2007.12.013
Falck J, Petrini JH, Williams BR, Lukas J, Bartek J (2002) The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat Genet 30(3):290–294. doi:10.1038/ng845
Lee J, Kumagai A, Dunphy WG (2001) Positive regulation of Wee1 by Chk1 and 14–3-3 proteins. Mol Biol Cell 12(3):551–563
Blasina A, de Weyer IV, Laus MC, Luyten WH, Parker AE, McGowan CH (1999) A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr Biol 9(1):1–10
Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP (2011) A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol 12(8):R78. doi:10.1186/gb-2011-12-8-r78
Sidorova JM, Breeden LL (1997) Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev 11(22):3032–3045
Sidorova JM, Breeden LL (2003) Rad53 checkpoint kinase phosphorylation site preference identified in the Swi6 protein of Saccharomyces cerevisiae. Mol Cell Biol 23(10):3405–3416
Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A (2007) Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell 131(5):915–926
Schwob E, Bohm T, Mendenhall MD, Nasmyth K (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79(2):233–244
Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ (1997) Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278(5337):455–460
Nyberg KA, Michelson RJ, Putnam CW, Weinert TA (2002) Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet 36:617–656
Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE (2000) Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res 60(21):5934–5936
Ahn JY, Li X, Davis HL, Canman CE (2002) Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem 277(22):19389–19395. doi:10.1074/jbc.M200822200
Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH (2006) Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J 25(13):3179–3190. doi:10.1038/sj.emboj.7601209
Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J (2000) Rapid destruction of human Cdc25A in response to DNA damage. Science 288(5470):1425–1429
Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J (2001) The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410(6830):842–847. doi:10.1038/35071124
Hoffmann I, Draetta G, Karsenti E (1994) Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J 13(18):4302–4310
Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281(5383):1677–1679
Dumaz N, Meek DW (1999) Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J 18(24):7002–7010. doi:10.1093/emboj/18.24.7002
Chehab NH, Malikzay A, Appel M, Halazonetis TD (2000) Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 14(3):278–288
Shieh SY, Ahn J, Tamai K, Taya Y, Prives C (2000) The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 14(3):289–300
Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387(6630):296–299. doi:10.1038/387296a0
Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW (2000) DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287(5459):1824–1827
Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D (1999) Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA 96(26):14973–14977
Chen L, Gilkes DM, Pan Y, Lane WS, Chen J (2005) ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 24(19):3411–3422. doi:10.1038/sj.emboj.7600812
Zhang Y, Xiong Y (2001) A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292(5523):1910–1915. doi:10.1126/science.1058637
Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13(12):1501–1512
Ekholm SV, Reed SI (2000) Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol 12(6):676–684
Segurado M, Tercero JA (2009) The S-phase checkpoint: targeting the replication fork. Biol Cell 101(11):617–627. doi:10.1042/BC20090053
Zegerman P, Diffley JF (2009) DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 8(9):1077–1088
Myung K, Kolodner RD (2002) Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99(7):4500–4507
Puddu F, Piergiovanni G, Plevani P, Muzi-Falconi M (2011) Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase epsilon. PLoS Genet 7(3):e1002022. doi:10.1371/journal.pgen.1002022
Paulovich AG, Margulies RU, Garvik BM, Hartwell LH (1997) RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 145(1):45–62
Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412(6846):553–557
Tercero JA, Longhese MP, Diffley JF (2003) A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11(5):1323–1336
Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412(6846):557–561
Santocanale C, Diffley JF (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395(6702):615–618
Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature 395(6702):618–621
Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ (1994) The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev 8(20):2401–2415
Longhese MP, Neecke H, Paciotti V, Lucchini G, Plevani P (1996) The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res 24(18):3533–3537
Brush GS, Morrow DM, Hieter P, Kelly TJ (1996) The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA 93(26):15075–15080
Marini F, Pellicioli A, Paciotti V, Lucchini G, Plevani P, Stern DF, Foiani M (1997) A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J 16(3):639–650
Longhese MP, Fraschini R, Plevani P, Lucchini G (1996) Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol Cell Biol 16(7):3235–3244
Navas TA, Zhou Z, Elledge SJ (1995) DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80(1):29–39
Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL (2008) Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell 32(1):106–117
Labib K (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24(12):1208–1219. doi:10.1101/gad.1933010
Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP (2010) Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467(7314):479–483. doi:10.1038/nature09377
Zegerman P, Diffley JF (2010) Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467(7314):474–478. doi:10.1038/nature09373
Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445(7125):328–332
Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445(7125):281–285
Segurado M, Diffley JF (2008) Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22(13):1816–1827
Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D (2008) Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J 27(18):2400–2410
Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M (2005) Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17(1):153–159
Smolka MB, Albuquerque CP, Chen SH, Zhou H (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA 104(25):10364–10369
de la Torre Ruiz MA, Lowndes NF (2000) DUN1 defines one branch downstream of RAD53 for transcription and DNA damage repair in Saccharomyces cerevisiae. FEBS Lett 485(2–3):205–206
Zhou Z, Elledge SJ (1993) DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75(6):1119–1127
Chen SH, Smolka MB, Zhou H (2007) Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae. J Biol Chem 282(2):986–995
Huang M, Zhou Z, Elledge SJ (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94(5):595–605
Zhao X, Rothstein R (2002) The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA 99(6):3746–3751
Lee YD, Wang J, Stubbe J, Elledge SJ (2008) Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell 32(1):70–80
Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9(8):616–627
Zachos G, Rainey MD, Gillespie DA (2005) Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol Cell Biol 25(2):563–574. doi:10.1128/MCB.25.2.563-574.2005
Zachos G, Rainey MD, Gillespie DA (2003) Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J 22(3):713–723. doi:10.1093/emboj/cdg060
Trenz K, Smith E, Smith S, Costanzo V (2006) ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25(8):1764–1774. doi:10.1038/sj.emboj.7601045
Grallert B, Boye E (2008) The multiple facets of the intra-S checkpoint. Cell Cycle 7(15):2315–2320
Lee J, Kumagai A, Dunphy WG (2003) Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, Rad17. Mol Cell 11(2):329–340
Guo Z, Kumagai A, Wang SX, Dunphy WG (2000) Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev 14(21):2745–2756
Kumagai A, Dunphy WG (2000) Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell 6(4):839–849
Chini CC, Chen J (2003) Human claspin is required for replication checkpoint control. J Biol Chem 278(32):30057–30062. doi:10.1074/jbc.M301136200
Gotter AL, Suppa C, Emanuel BS (2007) Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol 366(1):36–52. doi:10.1016/j.jmb.2006.10.097
Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK (2007) The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol 27(8):3131–3142. doi:10.1128/MCB.02190-06
Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A (2005) Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol 25(8):3109–3116. doi:10.1128/MCB.25.8.3109-3116.2005
Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang TH, Cordeiro-Stone M, Kaufmann WK, Abraham RT, Sancar A, Unsal-Kacmaz K (2010) Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem 285(22):16562–16571. doi:10.1074/jbc.M110.110304
Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ (1997) Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277(5331):1497–1501
Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H (1997) Mitotic and G2 checkpoint control: regulation of 14–3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277(5331):1501–1505
Sorensen CS, Syljuasen RG, FalckSchroeder J, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J (2003) Chk1 regulates the S phase checkpoint by coupling the physiological turnover, ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3(3):247–258
Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C (2006) The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J Biol Chem 281(41):30631–30644. doi:10.1074/jbc.M602982200
Kim ST, Xu B, Kastan MB (2002) Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev 16(5):560–570. doi:10.1101/gad.970602
Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J (2002) SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev 16(5):571–582. doi:10.1101/gad.970702
Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB (2004) Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev 18(12):1423–1438. doi:10.1101/gad.1200304
Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J (2008) Regulation of intra-S phase checkpoint by ionizing radiation (IR)-dependent and IR-independent phosphorylation of SMC3. J Biol Chem 283(28):19176–19183. doi:10.1074/jbc.M802299200
Norbury C, Blow J, Nurse P (1991) Regulatory phosphorylation of the p34cdc2 protein kinase in vertebrates. EMBO J 10(11):3321–3329
Krek W, Nigg EA (1991) Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: identification of major phosphorylation sites. EMBO J 10(2):305–316
Gould KL, Nurse P (1989) Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342(6245):39–45. doi:10.1038/342039a0
Lee MS, Enoch T, Piwnica-Worms H (1994) mik1 + encodes a tyrosine kinase that phosphorylates p34cdc2 on tyrosine 15. J Biol Chem 269(48):30530–30537
Parker LL, Atherton-Fessler S, Piwnica-Worms H (1992) p107wee1 is a dual-specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc Natl Acad Sci USA 89(7):2917–2921
Den Haese GJ, Walworth N, Carr AM, Gould KL (1995) The Wee1 protein kinase regulates T14 phosphorylation of fission yeast Cdc2. Mol Biol Cell 6(4):371–385
Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85. doi:10.1146/annurev.biochem.73.011303.073723
Raleigh JM, O’Connell MJ (2000) The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. J Cell Sci 113(Pt 10):1727–1736
O’Connell MJ, Raleigh JM, Verkade HM, Nurse P (1997) Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J 16(3):545–554
Rhind N, Furnari B, Russell P (1997) Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev 11(4):504–511
Zeng Y, Piwnica-Worms H (1999) DNA damage and replication checkpoints in fission yeast require nuclear exclusion of the Cdc25 phosphatase via 14–3-3 binding. Mol Cell Biol 19(11):7410–7419
Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T (1998) Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 395(6701):507–510
Lopez-Girona A, Furnari B, Mondesert O, Russell P (1999) Nuclear localization of Cdc25 is regulated by DNA damage and a 14–3-3 protein. Nature 397(6715):172–175
Amon A, Surana U, Muroff I, Nasmyth K (1992) Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature 355(6358):368–371
Sorger PK, Murray AW (1992) S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature 355(6358):365–368
Cohen-Fix O, Koshland D (1997) The metaphase-to-anaphase transition: avoiding a mid-life crisis. Curr Opin Cell Biol 9(6):800–806
Gardner R, Putnam CW, Weinert T (1999) RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J 18(11):3173–3185
Liang F, Wang Y (2007) DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol Cell Biol 27(14):5067–5078
Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K (1998) An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93(6):1067–1076
Wang H, Liu D, Wang Y, Qin J, Elledge SJ (2001) Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function. Genes Dev 15(11):1361–1372
Agarwal R, Tang Z, Yu H, Cohen-Fix O (2003) Two distinct pathways for inhibiting pds1 ubiquitination in response to DNA damage. J Biol Chem 278(45):45027–45033
Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO (1998) The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol 8(9):497–507
Shirayama M, Zachariae W, Ciosk R, Nasmyth K (1998) The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J 17(5):1336–1349
Schleker T, Shimada K, Sack R, Pike BL, Gasser SM (2010) Cell cycle-dependent phosphorylation of Rad53 kinase by Cdc5 and Cdc28 modulates checkpoint adaptation. Cell Cycle 9(2):350–363
Vidanes GM, Sweeney FD, Galicia S, Cheung S, Doyle JP, Durocher D, Toczyski DP (2010) CDC5 inhibits the hyperphosphorylation of the checkpoint kinase Rad53, leading to checkpoint adaptation. PLoS Biol 8(1):e1000286
Donnianni RA, Ferrari M, Lazzaro F, Clerici M, Tamilselvan Nachimuthu B, Plevani P, Muzi-Falconi M, Pellicioli A (2010) Elevated levels of the polo kinase Cdc5 override the Mec1/ATR checkpoint in budding yeast by acting at different steps of the signaling pathway. PLoS Genet 6(1):e1000763
Cheng L, Hunke L, Hardy CF (1998) Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol Cell Biol 18(12):7360–7370
Ferreira MG, Cooper JP (2004) Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev 18(18):2249–2254
Zhang Y, Shim EY, Davis M, Lee SE (2009) Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair (Amst) 8(10):1235–1241
Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y (2010) ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal 3(151):rs3. doi:10.1126/scisignal.2001034
Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics 9(6):1314–1323. doi:10.1074/mcp.M900616-MCP200
van Kouwenhove M, Kedde M, Agami R (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11(9):644–656. doi:10.1038/nrc3107
Wouters MD, van Gent DC, Hoeijmakers JH, Pothof J (2011) MicroRNAs, the DNA damage response and cancer. Mutat Res. doi:10.1016/j.mrfmmm.2011.03.012
Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T (2007) Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39(5):673–677. doi:10.1038/ng2003
Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Persengiev SP (2009) MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J 28(14):2090–2099. doi:10.1038/emboj.2009.156
Bu Y, Lu C, Bian C, Wang J, Li J, Zhang B, Li Z, Brewer G, Zhao RC (2009) Knockdown of Dicer in MCF-7 human breast carcinoma cells results in G1 arrest and increased sensitivity to cisplatin. Oncol Rep 21(1):13–17
Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, Chowdhury D (2011) miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 41(2):210–220. doi:10.1016/j.molcel.2010.12.005
Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456(7221):524–528
Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A (2008) 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456(7221):529–533
Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12(2):177–184. doi:10.1038/ncb2017
Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477(7363):179–184. doi:10.1038/nature10371
Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S (2009) Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460(7259):1132–1135. doi:10.1038/nature08235
Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460(7259):1136–1139. doi:10.1038/nature08290
Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460(7259):1140–1144. doi:10.1038/nature08311
Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K (2009) Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460(7259):1145–1148. doi:10.1038/nature08285
Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460(7259):1149–1153. doi:10.1038/nature08287
Andang M, Hjerling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, Pozas E, Bryja V, Halliez S, Nishimaru H, Wilbertz J, Arenas E, Koltzenburg M, Charnay P, El Manira A, Ibanez CF, Ernfors P (2008) Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature 451(7177):460–464. doi:10.1038/nature06488
Acknowledgments
We apologise to authors whose publications have not been cited due to space limitations. We thank T. Weinert for critical reading of the manuscript. This work was supported by an Irish Research Council for Science, Engineering and Technology (IRCSET) grant to K.F., a Cancer Research Ireland project grant to M.G. (CR105GRE), the European Union FP6 Integrated Project “Radiosensitivity of Individuals and Susceptibility to Cancer induced by Ionising RADiations” contract number F16R-CT-2003-508842 and Science Foundation Ireland Principal Investigator award 07/IN1/B958 to N.F.L.
Conflict of interest
All co-authors have seen and agree with the contents of the manuscript. The authors certify that the submission is original work and is not under review at any other publication. The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Finn, K., Lowndes, N.F. & Grenon, M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell. Mol. Life Sci. 69, 1447–1473 (2012). https://doi.org/10.1007/s00018-011-0875-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-011-0875-3