Abstract
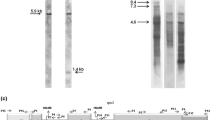
The organization of the genesnad3 andrps12 has been investigated in the mitochondrial genome of two dicotyledonous plants —Helianthus andMagnolia — and one monocotyledonous plant (Allium). These plants all contain a completerps12 gene downstream of thenad3 gene. This arrangement is thus highly conserved within angiosperms. The two genes are co-transcribed and the transcript is modified at several positions by RNA editing of the C to U-type, thus confirming that both genes encode functional proteins. Some 26, 35 and 27 editing events have been identified in the PCR-derivednad3-rps12 cDNA population from sunflower,Magnolia and onion, respectively. Editing of thenad3-rps12 transcript is thus more extensive inMagnolia than in the other angiosperms so far investigated and radically changes the genomically encoded polypeptide sequence. A novel species-specific codon modification was observed inMagnolia. Several homologous sites show differences in editing pattern among plant species. A C-to-U alteration is also found in the non-coding region separating thenad3 andrps12 genes in sunflower. The PCR-derived cDNA populations from thenad3-rps12 loci analysed were found to be differently edited. In addition the plant species show marked variations in the completeness of RNA editing, with only theMagnolia nad3 mRNA being edited fully.
Similar content being viewed by others
References
Anderson S, Bankier AT, Barrell BG, De Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Bland MM, Levings CS III, Matzinger DF (1986) The tobacco mitochondrial ATPase subunit 9 gene is closely linked to an open reading frame for a ribosomal protein. Mol Gen Genet 204:8–16
Bock H, Brennicke A, Schuster W (1994)Rps3 andrpl16 genes do not overlap inOenothera mitochondria: GTG as a potential translation initiation codon in plant mitochondria?. Plant Mol Biol 24:811–818
Bonnard G, Gualberto JM, Lamattina L, Grienenberger JM (1992) RNA editing in plant mitochondria. Crit Rev Plant Sci 10:503–524
Brennicke A, Grohmann L, Hiesel R, Knoop V, Schuster W (1993) The mitochondrial genome on its way to the nucleus: different stages of gene transfer in higher plants. FEBS Lett 325:140–145
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Conley CA, Hanson MR (1994) Tissue-specific protein expression in plant mitochondria. Plant Cell 6:85–91
Covello PS, Gray MW (1992) Silent mitochondrial and active nuclear genes for subunit 2 of cytochromec oxidase (cox2) in soybean: evidence for RNA-mediated gene transfer. EMBO J 11:3815–3820
Cronquist A (1988) The evolution and classification of flowering plants (2nd edn). New York Botanical Garden, New York
Cummings DJ, McNally KL, Domenico JM, Matsura ET (1990) The complete DNA sequence of the mitochondrial genome ofPodospora anserina. Curr Genet 17:375–402
Gray MW (1989) Origin and evolution of mitochondrial DNA. Annu Rev Cell Biol 5:25–50
Gray MW, Covello PS (1993) RNA editing in plant mitochondria and chloroplasts. FASEB J 7:64–71
Grohmann L, Brennicke A, Schuster W (1992) The mitochondrial gene encoding ribosomal protein S12 has been translocated to the nuclear genome inOenothera. Nucleic Acids Res 20:5641–5646
Grohmann L, Brennicke A, Schuster W (1993) Genes for mitochondrial ribosomal proteins in plants. In: Nierhaus KH, Franceschi F, Subramanian AR, Erdmann VA, Wittmann-Liebold B (eds) The translational apparatus, Plenum Press, New York, pp 599–607
Grohmann L, Thieck O, Herz U, Schroeder W, Brennicke A (1994) Translation ofnad9 mRNAs in mitochondria fromSolanum tuberosum is restricted to completely edited transcripts. Nucleic Acids Res 22:3304–3311
Gualberto JM, Wintz H, Weil JH, Grienenberger JM (1988) The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet 215:118–127
Gualberto JM, Bonnard G, Lamattina L, Grienenberger JM (1991) Expression of the wheat mitochondrialnad3-rps12 transcription unit: correlation between editing and mRNA maturation. Plant Cell 3:1109–1120
Hiesel R, von Haeseler A, Brennicke A (1994) Plant mitochondrial nucleic acid sequences as a tool for phylogenetic analysis. Proc Natl Acad Sci USA 91:634–638
Hunt MD, Newton KJ (1991) TheNCS3 mutation: genetic evidence for the expression of ribosomal protein genes inZea mays mitochondria.EMBO J 10:1045–1052
Karpinska B, Karpinski S, Hallgren JE (1995) The genes encoding subunit 3 of NADH dehydrogenase and ribosomal protein S12 are co-transcribed and edited inPinus sylvestris (L.) mitochondria. Curr Genet 28:423–428
Kim K-S, Schuster W, Brennicke A, Choi K-T (1991) Korean ginseng mitochondrial DNA encodes an intactrps12 gene downstream of thenad3 gene. Plant Physiol 97:1602–1603
Knoop V, Ehrhardt T, Lattig K, Brennicke A (1995) The gene for ribosomal protein S10 is present in mitochondria of pea and potato but absent from those ofArabidopsis andOenothera. Curr Genet 27:559–564
Lu B, Hanson MR (1992) A single nuclear gene specifies the abundance and extent of RNA editing of a plant mitochondrial transcript. Nucleic Acids Res 20:5699–5703
Lu B, Hanson MR (1994) A single homogeneous form of ATP6 protein accumulates inPetunia mitochondria despite the presence of differentially editedatp6 transcripts. Plant Cell 6:1955–1968
Moneger F, Smart CJ, Leaver CJ (1994) Nuclear restoration of cytoplasmic male sterility in sunflower is associated with the tissue-specific regulation of a novel mitochondrial gene. EMBO J 13:8–17
Nakazono M, Itadani H, Wakasugi T, Tsutsumi N, Sugiura M, Hirai A (1995) Therps3-rpl16-nad3-rps12 gene cluster in rice mitochondrial DNA is transcribed from alternative promoters. Curr Genet 27:184–189
Nugent JM, Palmer JD (1991) RNA-mediated transfer of the genecox2 from the mitochondrion to the nucleus during flowering plant evolution. Cell 66:473–481
Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S, Inokuchi H, Ozeki H (1986) Chloroplast gene organization deduced from the complete sequence of liverwortMarchantia polymorpha chloroplast DNA. Nature 322:572–574
Post LE, Nomura M (1980) DNA sequences from thestr operon ofE. coli. J Biol Chem 255:4660–4666
Quagliariello C, Saiardi A, Gallerani R (1990) The cytocrome oxidase subunit III gene in sunflower mitochondria is cotranscribed with an open reading frame conserved in higher plants. Curr Genet 18:355–363
Rasmussen J, Hanson MR (1989) A NADH dehydrogenase subunit gene is co-transcribed with the abnormalPetunia mitochondrial gene associated with cytoplasmic male sterility. Mol Gen Genet 215:332–336
Rickwood D, Wilson MT, Darley-Usmar VM (1987) Isolation and characteristics of intact mitochondria. In: Darley-Usmar VM, Rickwood D, Wilson MT (eds) Mitochondria. IRL Press, Oxford-Washington DC, pp 1–16
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual (2nd edn). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Schuster W (1993) Ribosomal protein generpl5 is cotranscribed with thenad3 gene inOenothera mitochondria. Mol Gen Genet 240:445–449
Schuster W, Brennicke A (1994) The plant mitochondrial genome: physical structure, information content, RNA editing, and gene migration to the nucleus. Annu Rev Plant Physiol Plant Mol Biol 45:61–78
Schuster W, Unseld M, Wissinger B, Brennicke A (1990a) Ribosomal protein S14 transcripts are edited inOenothera mitochondria. Nucleic Acids Res 18:229–233
Schuster W, Wissinger B, Unseld M, Brennicke A (1990b) Transcripts of the NADH-dehydrogenase subunit 3 gene are differentially edited inOenothera mitochondria. EMBO J 9:263–269
Siculella L, Palmer JD (1988) Physical and gene organization of mitochondrial DNA in fertile and male sterile sunflower. CMS-associated alterations in structure and transcription of theatpA gene. Nucleic Acids Res 16:3787–3799
Sutton CA, Conklin PL, Pruitt KD, Hanson MR (1991) Editing of pre-mRNA can occur beforecis- andtrans-splicing inPetunia mitochondria. Mol Cell Biol 11:4274–4277
Suzuki T, Kazama S, Hirai A, Akihama T, Kadowaki K (1991) The rice mitochondrialnad3 gene has an extended reading frame at its 5′ end: nucleotide sequence analysis of ricetrnS, nad3 andrps12 genes. Curr Genet 20:331–337
Takemura M, Oda K, Yamato K, Ohta E, Nakamura Y, Nozato N, Akashi K, Ohyama K (1992) Gene clusters for ribosomal proteins in the mitochondrial genome of a liverwort,Marchantia polymorpha. Nucleic Acids Res 20:3199–3205
Wahleithner JA, Wolstenholme DR (1988) Ribosomal protein S14 genes in broad bean mitochondrial DNA. Nucleic Acids Res 16:6897–6913
Wissinger B, Schuster W, Brennicke A (1990) Species-specific RNA editing patterns in the mitochondrialrps13 transcripts ofOenothera andDaucus. Mol Gen Genet 224:389–395
Yang AJ, Mulligan RM (1991) RNA editing intermediates ofcox2 transcripts in maize mitochondira. Mol Cell Biol 11:4278–4281
Ye F, Bernhardt J, Abel WO (1993) Genes for ribosomal proteins S3, L16, L5 and S14 are clustered in the mitochondrial genome ofBrassica napus L. Curr Genet 24:323–329
Zanlungo S, Quinones V, Moenne A, Holuigue L, Jordana X (1994) A ribosomal protein S10 gene is found in the mitochondrial genome inSolanum tuberosum. Plant Mol Biol 25:743–749
Author information
Authors and Affiliations
Additional information
Communicated by R. Hagemann
Rights and permissions
About this article
Cite this article
Perrotta, G., Regina, T.M.R., Quagliariello, C. et al. Conservation of the organization of the mitochondrialnad3 andrps12 genes in evolutionarily distant angiosperms. Molec. Gen. Genet. 251, 326–337 (1996). https://doi.org/10.1007/BF02172523
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02172523