Abstract
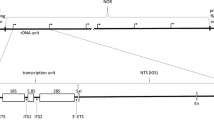
Nucleolar matrix structures were obtained under different extraction conditions from highly purified isolated nucleoli. Their ultrastructural appearance, protein composition and capacity to bind rDNA preferentially were studied in a model binding system. A region spanning approximately 25 kb in the rat ribosomal gene locus was screened for DNA sites capable of specifically interacting with the proteins of the nucleolar matrix (MARs). Two such sites were identified: one is located on an EcoRV-KpnI fragment in the 5′-nontranscribed spacer region, between two repetitive elements and close to the transcription initiation site; the other MAR is on a PvuII-BamHI fragment located in the 3′-nontranscribed region, encompassing an element 85% homologous to a B2-sequence. The two MARs are located in regions rich in polypyrimidine/polypurine tracks and contain a few elements homologous to the consensus sequence for topoisomerase II. This indicates that the “attachment sites” for the ribosomal genes belong to the same class of sequences as the MARs attaching the chromosomal DNA to the nuclear matrix.
Similar content being viewed by others
References
AdolphKW (1980) Organization of chromosomes in HeLa cells; Isolation of histone-depleted nuclei and nuclear scaffolds. J Cell Sci 42:291–304
AgutterPS (1972) The isolation of the envelopes from rat liver nuclei. Biochim Biophys Acta 255:394–401
ArisJB, BlobelG (1988) Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol 107:17–31
AvramovaZ, MikhailovI, TsanevR (1988) Metabolic behavior of a stable DNA-protein complex. Int J Biochem 20:61–65
BerezneyR, CoffeyD (1974) Identification of a nuclear protein matrix. Biochem Biophys Res Commun 60: 1410–1417
BerezneyR, CoffeyDS (1977) Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol 73:616–637
BodeJ, KohwiY, DickinsonL, JohT, KlehrD, MielkeC, Kohwi-ShigematsuT (1992) Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science 255:195–197
BodnarJW (1988) A domain model for eukaryotic DNA organization: a molecular basis for cell differentiation and chromosome evolution. J Theor Biol 132:479–507
BollaRI, BraatenDC, ShiomiY, HerbertMB, SchlessingerD (1985) Location of specific rDNA spacer sequences to the mouse L-cell nucleolar matrix. Mol Cell Biol 6:1287–1294
BourgeoisCA, BouvierD, SeveAP, HubertJ (1987) Evidence for the existence of a nuclear skeleton attached to the porecomplex lamina in human fibroblasts. Chromosoma 95:315–323
BraatenDC, ThomasJR, LittleRD, DicksonKR, GoldbergI, SchlessingerD, CiccodiolaA, D'UrsoM (1988) Locations and context of sequences that hybridize to poly(dG-dT), (dC-dA) in mammalian ribosomal DNAs and two X-linked genes. Nucleic Acids Res 16:865–881
BragaEA, AvdoninaTA, ZhurkinVB, NosikovVV (1985) Structural organization of rat ribosomal RNA genes: interspersed sequences and their putative role in the alignment of nucleosomes. Gene 36:249–262
CockerillPN, GarrardWT (1986) Chromosomal loop anchorage of the k-immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites Cell 44:273–282
CulottaV, Solner-WebbB (1988) Sites of topoisomerase I action on X. laevis ribosomal chromatin: transcriptionally active rDNA has an 200 bp repeating structure. Cell 52:585–597
DavisAH, ReudelhuberTL, GarrardWT (1983) Variegated chromatin structures of mouse ribosomal RNA genes. J Mol Biol 167:133–155
DickinsonLA, JohT, KohwiY, Kohwi-ShigematsuT (1992) A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70:631–645
FeyEG, KrochmalnicG, PennmanS (1986) The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and (RNP)-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol 102:1654–1665
FrankeWW, KleinschmidtJA, SpringH, KrohneG, GrundC, TrendelenburgMF, StoehrN, ScheerU (1981) A nucleolar skeleton of protein filaments demonstrated in amplified nucleoli of X. laevis. J Cell Biol 90:289–299
GajdardjievaKC, MarkovDV, DimovaRN, KermekchievMB, TodorovIT, DabevaMD, HadjiolovAA (1982) Isolation and initial characterization of nuclear fibrillar remnants from the liver of rats treated with d-galactozamin. Exp Cell Res 140:95–104
GasserSM, LaemmliUK (1986) The organization of chromatin loops: characterization of a scaffold attachment site. EMBO J 5:511–517
GoldmanMA (1988) The chromatin domain as a unit of gene regulation. BioEssays 9:50–55
GrossDS, GarrardWT (1987) Poising chromatin for transcription. Trends Biochem Sci 12:293–297
JacksonDA, McCreadySJ, CookPR (1981) RNA is synthesised at the nuclear cage. Nature 292:552–555
JacksonDA, CookPR, PatelSB (1984) Attachment of repeated sequences to the nuclear cage. Nucleic Acids Res 12:6709–6726
JamesGT, YeomanLC, MatsuiS, GoldbergAH, BuschH (1977) Isolation and characterization of nonhistone chromosomal protein C-14 which stimulates RNA synthesis. Biochemistry 16:2384–2389
JordanEG (1984) Nucleolar nomenclature. J Cell Sci 67:217–220
KasE, LaemmliUK (1992) In vivo topoisomerase II cleavage of Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J 11:705–716
KaufmannSH, CoffeyDS, ShaperJH (1981) Considerations in the isolation of rat liver nuclear matrix, nuclear envelope and pore-complex lamina. Exp Cell Res 132:105–121
KeppelF (1986) Transcribed human ribosomal RNA genes are attached to the nuclear matrix. J Mol Biol 187:15–21
LaemmliUK (1972) Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 227:680–685
MarilleyM, Gassend-BonnetG (1989) Supercoiled loop organization of genomic DNA: a close relationship between loop domains expression units and replicon organization in rDNA from X. laevis. Exp Cell Res 180:475–489
MillerOL, BakkenAH (1972) Morphological studies on transcription. Acta Endocrinol [Suppl] 168:155–173
MillerTE, HuangCY, PogoAO (1978) Rat liver nuclear skeleton and ribonucleoprotein complexes containing hnRNA. J Cell Biol 76:675–691
MirskyAE, RisH (1951) The composition and structure of isolated chromosomes. J Gen Physiol 34:475–492
MountyKL, DounceAL (1958) The properties and the enzymatic degradation of deoxyribonucleoprotein from liver cell nuclei. J Gen Physiol 41:595–608
MullerMT, PfundWP, MehtaVP, TraskDK (1985) Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J 4:1237–1243
OchsRL, SmetanaK (1991) Detection of fibrillarin in nucleolar remnants and the nucleolar matrix. Exp Cell Res 197:183–190
OlsonMOJ, WallaceMO, HerreraAH, Marshal-CarlsonL, HuntRC (1986) Preribosomal ribonucleoprotein particles are a major component of a nucleolar matrix fraction. Biochemistry 25:484–491
PardollDM, VogelsteinB (1980) Sequence analysis of nuclear matrix associated DNA from rat liver. Exp Cell Res 128:466–470
RoseKM, SzopaJ, HanF-S, ChengY-C, RichterA, ScheerU (1988) Association of DNA topoisomerase I and polymerase I: a possible role for topoisomerase I in ribosomal gene transcription. Chromosoma 96:411–416
RothblumLI, MamrakPM, KunkleHM, OlsonOJ, BushH (1977) Fractionation of nucleoli. Enzymatic and two-dimensional polyacrylamide gel electrophoresis. Biochemistry 16:4716–4721
ShiomiY, PoweresJ, BollaRI, NguyenTV, SchlessingerD (1986) Proteins and RNA in mouse L cell core nucleoli and nucleolar matrix. Biochemistry 25:5745–5751
SmithHC, RothblumLI (1987) Ribosomal DNA sequences attached to the nuclear matrix. Biochem Genet 25:863–879
SmithHC, OchsRL, LinD, ChinaultAC (1987) Ultrastructural and biochemical comparison of nuclear matrices prepared by high salt or LIS extraction. Mol Cell Biochem 77:49–61
StephanovaE (1989) A method for the isolation of nucleoli from mouse liver. C R Acad Sci Bulg 42:123–126
StephanovaE, ValkovN (1990) Rat liver nucleoli: characterization of the residual skeletal structure. C R Acad Sci Bulg 43:87–90
StephanovaE, ValkovN (1991) Rat liver nucleoli: biochemical characterization of the residual skeletal structure. C R Acad Sci Bulg 44:63–66
ThomasJR, BollaRI, RumbyrtJS, SchlessingerD (1985) DNase I resistant nontranscribed spacer segments of mouse ribosomal DNA contain poly(gD-dI), poly(dA-dC). Proc Natl Acad Sci USA 82:7595–7598
TsanevR, AvramovaZ (1981) Nonprotamine nucleoprotein ultrastructures in mature ram sperm nuclei. Eur J Cell Biol 24:139–145
VerheijenR, van VenrooijW, RamaekersF (1988) The nuclear matrix: structure and composition. J Cell Sci 90:11–36
YavachevLP, GeorgievOI, BragaEA, AvdoninaTA, ZhurkinVB, NosikovVV, HadjiolovAA (1986) Nucleotide sequence analysis of the spacer regions flanking the rat rRNA transcript unit and identification of repetitive elements. Nucleic Acids Res 14:2799–2810
ZhangH, WangJ, LiuLF (1988) Involvement of topoisomerase i in transcription of human ribosomal RNA genes. Proc Natl Acad Sci USA 85:1060–1064
Author information
Authors and Affiliations
Additional information
Communicated by: W.C. Earnshaw
Rights and permissions
About this article
Cite this article
Stephanova, E., Stancheva, R. & Avramova, Z. Binding of sequences from the 5′- and 3′-nontranscribed spacers of the rat rDNA locus to the nucleolar matrix. Chromosoma 102, 287–295 (1993). https://doi.org/10.1007/BF00352403
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00352403