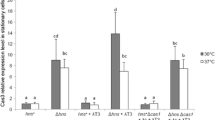
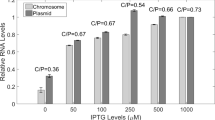
Summary
A mutation (dicA1) of a repressor gene located in the terminus region of the Escherichia coli chromosome has previously been shown to lead to temperature-dependent inhibition of division, and to be complemented by plasmids carrying either dicA or an adjacent gene dicC. In this study, operon fusions in the region coding for the division inhibition gene dicB have been used to show that temperature sensitivity does not result from high temperature inactivation of the dicA repressor. Sequence comparisons indicate that dicA and dicC are similar to genes c2 and cro respectively of bacteriophage P22, and carry similarly organized tandem operators, indicating a common evolutionary origin for dicAC and P22 immC. Nevertheless, the consensus half-operator sequence of dicAC, TGTTAGYYA, differs significantly from that of P22 immC (ATTTAAGAN). an analysis of the in vivo control of promoters dicAp, dicBp and dicCp placed upstream of malQ shows that the dicAC system is functionally similar to that of an immunity region, with the possible exception of an absence of pairwise cooperative binding. Our results also indicate that the dicA1 mutation causes a switch to permanent control by dicC at all temperatures.
Similar content being viewed by others
References
Anderson JE, Ptashne M, Harrison SC (1984) Cocrystals of the DNA-binding domain of phage 434 repressor and a synthetic phage 434 operator. Proc Natl Acad Sci USA 81:1307–1311
Anderson JE, Ptashne M, Harrison SC (1985) A phage repressoroperator complex at Å resolution. Nature 316:596–601
Backhaus H, Petri JB (1984) Sequence analysis of a region from the early right operon in phage P22 including the replication genes 18 and 12. Gene 32:289–303
Bech FW, Jorgensen ST, Diderischen B, Karlström OH (1985) Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J 4:1059–1065
Béjar S, Bouché JP (1985) A new dispensable genetic locus of the terminus region involved in control of cell division in Escherichia coli. Mol Gen Genet 201:146–150
Béjar S, Cam K, Bouché JP (1986) Control of cell division in Escherichia coli. DNA sequence of dicA and of a second gene complementing mutation dicA1, dicC. Nucleic Acids Res 14:6821–6833
Brody H, Greener A, Hill CW (1985) Excision and reintegration of the Escherichia coli K12 chromosomal element e14. J Bacteriol 161:1112–1117
Churchward G, Belin D, Nagamine Y (1984) A pSC101-derived plasmid which shows no homology with other commonly used cloning vectors. Gene 31:165–171
Craig N, Nash H (1984) Escherichia coli integration host factor binds to specific sites in DNA. Cell 39:707–716
Dayhoff MO, schwartz RM, Orcutt BC (1978) In: Dayhoff MO (ed) Anas of protein sequence and structure, vol. 5, suppl. 3. National Biomedical Research Foundation, Silver Spring, MD
Feinstein SI, Low KB (1982) Zygotic induction of the rac locus can cause cell death in Escherichia coli. Mol Gen Genet 187:231–255
Franklin NC (1985a) Conservation of genome but not sequence in the transcription antitermination determinants of bacteriophages λ, ø 21 and P22. J Mol Biol 181:75–84
Franklin NC (1985b) “N” transcription antitermination proteins of bacteriophages λ, ø 21 and P22. J Mol Biol 181:85–91
Gamas P, Chandler MG, Prentki P, Galas DJ (1987) Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hotspot in pBR322. J Mol Biol 195:261–272
Henson JM, Kuempel PL (1985) Deletion of the terminus region (340 kilobase pairs of DNA) from the chromosome of Escherichia coli. Proc Natl Acad Sci USA 82:3766–3770
Hochschild A, Irwin N, Ptashne M (1983) Repressor structure and the mechanism of positive control. Cell 32:319–325
Johnson AD, Meyer BJ, Ptashne M (1978) Mechanism of action of the Cro protein of bacteriophage λ. Proc Natl Acad Sci USA 75:1783–1787
Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GR, Ptashne M (1981). λ repressor and Cro, components of an efficient molecular switch. Nature 294:217–223
Kanehisha M (1984) Use of statistical criteria for screening potential homologies in nucleic acid sequences. Nucleic Acids Res 12:203–213
Kroos L, Kaiser D (1984) Construction of Tn5-lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci USA 81:5816–5820
Leong J, Nunes-Duby S, lesser C, Youderian P, Susskind M, Landy A (1985) The ø80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem 260:4468–4477
Low KB (1973) Restoration by the rac locus of recombinant forming ability in recB - and recC - merozygotes of Escherichia coli K12. Mol Gen Genet 122:119–130
Maguin E, Brody H, Hill CW, D'Ari R (1986) SOS-associated division inhibition gene sfiC is part of excisable element e14 of Escherichia coli. J Bacteriol 168:465–467
Meyer BM, Maurer R, Ptashne M (1980) Gene regulation at the right operator (O R) of bacteriophage λ. II. O R1, OR2 and O R3: their roles in mediating the effects of repressor and Cro. J Mol Biol 139:163–194
Miller J (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Pabo CO, Lewis M (1982) The operator-binding domain of λ repressor: structure and DNA recognition. Nature 298:443–447
Pabo CO, Sauer RT (1984) Protein-DNA recognition. Annu Rev Biochem 53:293–321
Pabo CO, Sauer RT, Sturtevant JM, Ptashne M (1979) The repressor contains two domains. Proc Natl Acad Sci USA 76:1608–1611
Poteete AR, Ptashne M (1982) Control of transcription by the bacteriophage P22 repressor. J Mol Biol 157:21–48
Poteete AR, Ptashne M, Ballivet M, Eisen H (1980) Operator sequences of bacteriophages P22 and ø21. J Mol Biol 137:81–91
Poteete AR, Hehir K, Sauer RT (1986) Bacteriophage P22 Cro protein: Sequence, purification and properties. Biochemistry 25:251–256
Prentki P, Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313
Ptashne M (1986) A genetic switch. Cell Press and Blackwell publications
Raibaud O, Schwartz M (1984) Positive control of transcription in bacteria. Annu Rev Genet 18:173–206
Raibaud O, Mock M, Schwartz M (1984) A technique to integrate any DNA fragment into the chromosome of Escherichia coli. Gene 29:231–241
Raibaud O, Guttierez C, Schwartz M (1985) Essential and nonessential sequences in malPp, a positively controlled promoter in Escherichia coli. J Bacteriol 161:1201–1208
Sauer RT, Pan J, Hopper P, Hehir K, Brown J, Poteete AR (1981) Primary structure of the phage P22 repressor and its gene c2. Biochemistry 20:3591–3598
Sauer RT, Ross MR, Ptashne M (1982a) Cleavage of the λ and P22 repressors by RecA protein. J Biol Chem 247:4458–4462
Sauer RT, Yocum RR, Doolitle RF, Lewis M, Pabo CO (1982b) Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature 298:447–451
Vidal-Ingiliardi D, Raibaud O (1985) A convenient technique to compare the efficiency of promoters in Escherichia coli. Nucleic Acids Res 13:5919–5926
Author information
Authors and Affiliations
Additional information
Communicated by R. Devoret
Rights and permissions
About this article
Cite this article
Béjar, S., Bouché, F. & Bouché, JP. Cell division inhibition gene dicB is regulated by a locus similar to lamboid bacteriophage immunity loci. Molec. Gen. Genet. 212, 11–19 (1988). https://doi.org/10.1007/BF00322439
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00322439