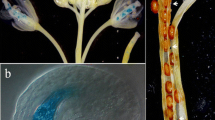
Abstract
Five constructions containing deletions of the promoter from an auxin-inducible gene of Arabidopsis thaliana, AtAux2-11, were fused to the coding region of the reporter gene LacZ, which encodes β-galactosidase, and a polyadenylation 3′-untranslated nopaline synthase sequence from Agrobacterium. These chimeric genes were introduced into Arabidopsis by Agrobacterium tumefaciens-mediated transformation, and expression of the gene was examined by spectrophotometric and histochemical analyses. A 600 bp fragment from the AtAux2-11 promoter conferred histochemical patterns of staining similar to the longest 5′ promoter tested, a 3.0 kb fragment. Localization of AtAux2-11/LacZ activity in the transgenic plants revealed spatial and temporal expression patterns that correlated with tissues and cells undergoing physiological processes modulated by auxin. LacZ activity was expressed in the elongating region of roots, etiolated hypocotyls, and anther filaments. Expression was detected in the vascular cylinder of the root and the vascular tissue, epidermis, and cortex of the hypocotyl, and filament. The AtAux2-11/LacZ gene was preferentially expressed in cells on the elongating side of hypocotyls undergoing gravitropic curvature. Expression of the chimeric gene in the hypocotyls of light-grown seedlings was less than that in etiolated seedling hypcotyls. The AtAux2-11/LacZ gene was active in the root cap, and expression in the root stele increased at sites of lateral root initiation. Staining was evident in cell types that develop lignified cell walls, e.g. trichomes, anther endothecial cells, and especially developing xylem. The chimeric gene was not expressed in primary meristems. While the magnitude of expression increased after application of exogenous auxin (2,4-D), the histochemical localization of AtAux2-11/LacZ remained unchanged.
Transgenic plants with a 600 bp promoter construct (−0.6 kb AtAux2-11/LacZ) had higher levels of basal and auxin-inducible expression than plants with a 3.0 kb promoter construct. Transgenic plants with a −500 bp promoter had levels of expression similar to the −3.0 kb construct. The −0.6 kb AtAux2-11/LacZ gene responded maximally to a concentration of 5 × 10−6 to 5 × 10−5 M 2,4-D and was responsive to as little as 5 × 10−8 M. The evidence presented here suggests that this gene may play a role in several auxin-mediated developmental and physiological processes.
Similar content being viewed by others
References
Ainley WM, Walker JC, Nagao RT, Key JL: Sequence and characterization of two auxin-regulated genes from soybean. J Biol Chem 263: 10658–10666 (1988).
Alliote T, Tiré C, Engler G, Peleman J, Caplan A, VanMontagu M, Inzé D: An auxin-regulated gene of Arabidopsis thaliana encodes a DNA-binding protein. Plant Physiol 89: 743–752 (1989).
An G, Mitra A, Choi H, Costa M, An K, Thornburg RW, Ryan C: Functional analyses of the 3′ control region of the potato wound-inducible protease inhibitor II gene. Plant Cell 1: 115–122 (1989).
Baulcombe DC, Key JL: Polyadenylated RNA sequences which are reduced in concentration following auxin treatment of soybean hypocotyls. J Biol Chem 255: 8907–8913 (1980).
Blakely LM, Blakely RM, Colowit PM, Elliot DS: Experimental studies on lateral root formation in radish seedling roots. II. Analysis of the dose-response to exogenous auxin. Plant Physiol 87: 414–419 (1988).
Conner TW, Goekjian VH, Lafayette PR, Key JL: Structure and expression of two auxin-inducible genes from Arabidopsis. Plant Mol Biol 15: 623–632 (1990).
Ditta G, Stanfield G, Corbin D, Helinski DR: Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA 77: 7347–7354 (1980).
Figurski DH, Helinski DR: Replication of an origin containing a derivative of plasmid RK2, dependence on a plasmid function provided in trans. Proc Natl Acad Sci USA 76: 1648–1652 (1979).
Gee MA, Hagen G, Guilfoyle TJ: Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3: 419–430 (1991).
Guilfoyle TJ: Auxin-regulated gene expression in higher plants. CRC Crit Rev Plant Sci 4: 247–276 (1986).
Hagen G, Kleinschmidt A, Guilfolye TJ: Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 162: 147–153 (1984).
Hagen G, Guilfoyle TJ: Rapid induction of selective transcription by auxins. Mol Cell Biol 5: 1197–1203 (1985).
Hood EE, Helmer GL, Fraley RT, Chilton M-D: The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of the T-DNA. J Bact 168: 1291–1298 (1986).
Jeffs RA, Northcote DH: The influence of indolyl-3-acetic acid and sugar on the pattern of induced differentiation in plant tissue culture. J Cell Sci 2: 77–88 (1967).
Key J, Kroner P, Walker J, Hong JC, Ulrich TH, Ainley WM, Gantt JS, Nagao RT: Auxin-regulated gene expression. Phil Trans R Soc Lond B 314: 427–440 (1986).
Laemmli UK: Cleavage of structural proteins during the assembly of bacteriophage T4. Nature 277: 680–685 (1970).
Li Y, Hagen G, Guilfoyle TJ: An auxin-responsive promoter is differentially induced by auxin gradients during tropisms. Plant Cell 3: 1167–1175 (1991).
Lyndon RF: Plant Development: The Cellular Basis. Unwin-Hyman, London (1990).
McClure BA, Guilfoyle T: Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 (1987).
Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1972).
Reddy ASN, Poovaiah BW: Molecular cloning and sequencing of a cDNA for an auxin-repressed mRNA: correlation between fruit growth and repression of the auxinregulated gene. Plant Mol Biol 14: 127–136 (1990).
Sachs T: The control of patterned differentiation of vascular tissues. Adv Bot Res 9: 151–262 (1981).
Sachs T: Axiality and polarity in plants. In: Barlow PW, Carr DJ (eds) Positional Controls in Plant Development, pp. 193–224. Cambridge University Press, Cambridge (1984).
Takahashi Y, Kuroda H, Tanaka T, Machida Y, Takabe I, Nagata T: Isolation of an auxin-regulated cDNA expressed during the transition from G0 to S phase in tobacco mesophyll protoplast. Proc Natl Acad Sci USA 86: 9279–9283 (1989).
Teeri TH, Lehväslaiho H, Franck M, Uotila J, Heino P, Palva ET, VanMontagu M, Herrera-Estrella L: Gene fusions to lacZ reveal new expression patterns of chimeric genes in transgenic plants. EMBO J 8: 343–350 (1989).
Theologis A, Huynh TV, Davis RW: Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol 3: 53–68 (1985).
Valvekens D, VanMontagu M, VanLijsebettens M: Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 (1980).
van derZaal EJ, Mennes AM, Libbenga KR. Auxin-induced rapid changes in translatable mRNAs in tobacco cell suspensions. Planta 172: 514–519 (1987).
Walker JC, Key JL: Isolation of cloned cDNAs to auxinresponsive poly(A)+ RNAs of elongating soybean hypocotyl. Proc Natl Acad Sci USA 79: 7185–7189 (1982).
Author information
Authors and Affiliations
Additional information
co-first authors
Rights and permissions
About this article
Cite this article
Wyatt, R.E., Ainley, W.M., Nagao, R.T. et al. Expression of the Arabidopsis AtAux2-11 auxin-responsive gene in transgenic plants. Plant Mol Biol 22, 731–749 (1993). https://doi.org/10.1007/BF00027361
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00027361