Abstract
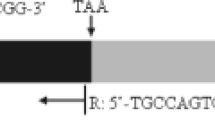
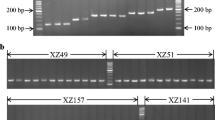
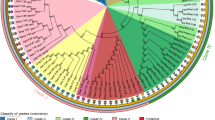
While the two amylase genes ofDrosophila melanogaster are intronless, the three genes ofD. pseudoobscura harbor a short intron. This raises the question of the common structure of theAmy gene in Drosophila species. We have investigated the presence or absence of an intron in the amylase genes of 150 species of Drosophilids. Using polymerase chain reaction (PCR), we have amplified a region that surrounds the intron site reported inD. pseudoobscura and a few other species. The results revealed that most species contain an intron, with a variable size ranging from 50 to 750 bp, although the very majoritary size was around 60–80 bp. Several species belonging to different lineages were found to lack an intron. This loss of intervening sequence was likely due to evolutionarily independent and rather frequent events. Some other species had both types of genes: In theobscura group, and to a lesser extent in theananassae subgroup, intronless copies had much diverged from intron-containing genes. Base composition of short introns was found to be variable and correlated with that of the surrounding exons, whereas long introns were all A-T rich. We have extended our study to non-Drosophilid insects. In species from other orders of Holometaboles, Lepidoptera and Hymenoptera, an intron was found at an identical position in theAmy gene, suggesting that the intron was ancestral.
Similar content being viewed by others
References
Balm E (1967) Crossing over in the chromosomal region determining amylase isozymes inDrosophila melanogaster. Hereditas 58:1–12
Bingham PM, Chou T-B, Mims I, Zachar Z (1988) On/off regulation of gene expression at the level of splicing. Trends Genet 4:134–138
Boer PH, Hickey DA (1986) The alpha-amylase gene inDrosophila melanogaster: nucleotide sequence, gene structure and expression motifs. Nucleic Acids Res 14:8399–8411
Bowtell DDL, Simon MA, Rubin GM (1988) Nucleotide sequence and structure of thesevenless gene ofDrosophila melanogaster. Genes Dev 2:620–634
Brown CJ, Aquadro CF, Anderson WW (1990) DNA sequence evolution of the amylase multigene family inDrosophila pseudoobscura. Genetics 126:131–138
Cariou M-L, Lachaise D, Sourdis J, Tsacas L, Krimbas C, Ashbumer M (1988) New African species in theDrosophila obscura species group: genetic variation, differentiation and evolution. Heredity 61: 73–84
Cavalier-Smith T (1991) Intron phylogeny: a new hypothesis. Trends Genet 7:145–148
Csank C, Taylor FM, Martindale DW (1990) Nuclear pre-mRNA introns: analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res 18: 5133–5141
Da Lage J-L, Carion M-L, David JR (1989) Geographical polymorphism of amylase inDrosophila ananassae and its relatives. Heredity 63:67–72
Da Lage J-L, Cariou M-L (1993) Organization and structure of the amylase gene family. In: Tobari YN (ed)Drosophila ananassae, genetical and biological aspects. Japan Scientific Societies Press, Karger, Tokyo, pp 171–181
Da Lage J-L, Lemeunier F, Cariou M-L, David JR (1992) Multiple amylase genes inDrosophila ananassae and related species. Genet Res Camb 59:85–92
Daïnou O, Cariou M-L, David JR, Hickey D (1987) Amylase gene duplication: an ancestral trait in theDrosophila melanogaster species subgroup. Heredity 59:245–251
Di Bello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM (1991) TheDrosophila Broad-Complex encodes a family of related proteins containing zinc-fingers. Genetics 129:385–397
Doolittle WF (1987) The origin and function of intervening sequences in DNA: a review. Am Nat 130:915–928
Ellsworth DL, Hewett-Emmett D, Li W-H (1994) Evolution of base composition in the insulin and insulin-like growth factor genes. Mol Biol Evol 11:875–885
Geiss KT, Abbas GM, Makaroff CA (1994) Intron loss from the NADH dehydrogenase subunit 4 gene of lettuce mitochondrial DNA: evidence for homologous recombination of a cDNA intermediate. Mol Gen Genet 243:97–105
Gemmill RM, Levy JN, Doane WW (1985) Molecular cloning of alpha-amylase genes fromDrosophila melanogaster. I Clone isolation by use of a mouse probe. Genetics 110:299–312
Gilbert W (1978) Why genes-in-pieces? Nature 271:501
Giroux MJ, Clancy M, Baier J, Ingham L, McCarthy D, Hannah LC (1994)De novo synthesis of an intron by the maize transposable elementDissociation. Proc Natl Acad Sci USA 91:12150–12154
Gloor G, Engels W (1991) Single fly DNA preps for PCR. Dros Inf Serv 74:148–149
Goodall GJ, Filipowicz W (1989) The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58:473–483
Gumucio DL, Wiebauer K, Caldwell RM, Samuelson L, Meisler MH (1988) Concerted evolution of human amylase genes. Mol Cell Biol 8:1197–1205
Guo M, Lo PCH, Mount SM (1993) Species-specific signals for the splicing of a short intron in vitro. Mol Cell Biol 13:1104–1118
Hawkins JD (1988) A survey on intron and exon lengths. Nucleic Acids Res 16:9893–9905
Heberlein U, Rubin G (1990) Structural and functional comparisons of theDrosophila virilis andDrosophila melanogaster rough genes. Proc Natl Acad Sci USA 87:5916–5920
Hickey DA (1979) The geographical pattern of an enzyme polymorphism inD. melanogaster. Genetica 51:1–4
Janecek S (1994) Sequence similarities and evolutionary relationships of microbial, plant and animal alpha-amylases. Eur J Biochem 224: 519–524
Kassis JA, Poole SJ, Wright DK, O'Farrell PH (1986) Sequence conservation in the protein coding and intron regions of theengrailed transcription unit. EMBO J 5:3583–3589
Keller EB, Noon WA (1985) Intron splicing: a conserved internal signal in introns of Drosophila pre-mRNAs. Nucleic Acids Res 13:4971–4981
Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS (1991) The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67:59–77
Kristensen NP (1991) Phylogeny of extant Hexapods. In: Nauman ID (ed) The insects of Australia. Carlton, Victoria, Melbourne, Univ. Press, pp 125–140
Kukalova-Peck J (1991) Fossil history and the evolution of Hexapod structures. In: Nauman ID (ed) The insects of Australia. Carlton, Victoria, Melbourne, Univ. Press, pp 141–179
Michael WM, Bowtell DDL, Rubin GM (1990) Comparison of the sevenless genes ofDrosophila virilis andDrosophila melanogaster. Proc Natl Acad Sci USA 87:5351–5353
Mount SM (1982) A catalogue of splice junction sequences. Nucleic Acids Res 10:459–472
Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C (1992) Splicing signals in Drosophila: inron size, information content, and consensus sequences. Nucleic Acids Res 20:4253–4262
Neufeld TP, Carthew RW, Rubin GM (1991) Evolution of gene position: chromosomal arrangement and sequence comparison of theDrosophila melanogaster andDrosophila virilis sina andRh4 genes. Proc Natl Acad Sci USA 88:10203–10207
Ohta T (1983) On the evolution of multigene families. Theor Popul Biol 23:216–240
Payant V, Abukashawa S, Sasseville M, Benkel BF, Hickey DA, David J (1988) Evolutionary conservation of the chromosomal configuration and regulation of amylase genes among eight species of theDrosophila melanogaster species subgroup. Mol Biol Evol 5:560–567
Pélandakis M, Higgins DG, Solignac M (1991) Molecular phylogeny of the subgenusSophophora ofDrosophila derived from large subunit of ribosomal RNA sequences. Genetica 84:87–94
Purugganan M, Wessler S (1993) The splicing of transposable elements and its role in intron evolution. In: McDonald JF (ed) Transposable elements and evolution. Kluwer Academic, Dordrecht, pp 28–36
Riley MA (1989) Nucleotide sequence of theXdh region inDrosophila pseudoobscura and an analysis of the evolution of synonymous codons. Mol Biol Evol 6:33–52
Rogers JH (1990) The role of introns in evolution. FEBS Lett 268: 339–343
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463
Sharp PA (1985) On the origin of RNA splicing and introns. Cell 42:397–400
Shibata H, Yamazaki T (1995) Molecular evolution of the duplicatedAmy locus inDrosophila melanogaster species subgroup: concerted evolution only in coding region and excess of nonsynonymous substitutions in speciation. Genetics 141:223–236
Stephan W, Rodriguez VS, Zhou B, Parsch J (1994) Molecular evolution of the metallothionein geneMtn in themelanogaster species group: results fromDrosophila ananassae. Genetics 138:135–143
Tadlaoui-Ouafi A (1993) Evolution structurale et moléculaire de la famille mulrigénique Amylase chez quelques Drosophilidae. Thesis, Université Pierre et Marie Curie, Paris, 142 pp
Talerico M, Berget SM (1994) Intron definition in splicing of smallDrosophila introns. Mol Cell Biol 14:3434–3445
Author information
Authors and Affiliations
Additional information
Correspondence to: J-L. Da Lage
Rights and permissions
About this article
Cite this article
Da Lage, JL., Wegnez, M. & Cariou, ML. Distribution and evolution of introns in drosophila amylase genes. J Mol Evol 43, 334–347 (1996). https://doi.org/10.1007/BF02339008
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02339008