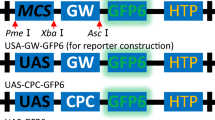
Abstract
The use of reporter genes to characterise sequence elements that act to regulate gene expression in transgenic plants has been vital to the development of foreign gene expression strategies for use in cereal transformation. ThegusA locus ofEscherichia coli, which encodes the enzymeβ-glucuronidase (GUS), is by far the most popular reporter gene used in plant transformation. In this paper we extend the utility of the GUS reporter gene system in cereal transformation by describing and evaluating a number of novel constructs suitable for use in direct gene transfer experiments. These plasmids are all available from the Molecular Genetic Resource Service of the Center for the Application of Molecular Biology to International Agriculture.
Similar content being viewed by others
References
Bower R, Birch RG: Transgenic sugarcane plants via microparticle bombardment. Plant J 2: 409–416 (1992).
Callis J, Fromm M, Walbot V: Introns increase gene expression in cultured maize cells. Genes Devel 1: 1183–1200 (1987).
Cao J, Duan X, McElroy D, Wu R: Regeneration of herbicide resistant transgenic rice plants following microparticle-mediated transformation of suspension culture cells. Plant Cell Rep 11: 586–591 (1992).
Chamberlain DA, Brettell RIS, Last DI, Witrzens B, McElroy D, Dennis ES: The use of the Emu promoter with antibiotic and herbicide resistance genes for the selection of transgenic wheat callus and rice plants. Aust J Plant Physiol 21: 95–112 (1994).
Christensen AH, Sharrock RA, Quail PH: Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18: 675–689 (1992).
Christou P, Ford T, Koffron M: Production of transgenic rice (Oryza sativa L.) plants from agronomically important indica and japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos. Bio/technology 9: 957–962 (1991).
Farrell LB, Beachy RN: Manipulation ofβ-glucuronidase for use as a reporter in vacuolar targeting studies. Plant Mol Biol 15: 821–825 (1990).
Finnegan EJ, Brettell RIS, Dennis ES: The role of DNA methylation in the regulation of plant gene expression. In: Jost JP, Saluz HP (eds) DNA Methylation: Molecular Biology and Biological Significance, pp. 218–261. Birkhäuser Verlag, Basel (1993).
Fromm ME, Morrish F, Armstrong C, Williams R, Thomas J, Klein TM: Inheritance and expression of chimaeric genes in the progeny of transgenic maize plants. Bio/technology 8: 833–844 (1990).
Gordon-Kamm WJ, Spencer TM, Mangano ML, Adams TR, Daines RJ, Start WG, O'Brien JV, Chambers SA, Adams WR, Willetts NG, Rice TB, Mackey CJ, Krueger RW, Kausch AP, Lemaux PG: Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell 2: 603–618 (1990).
Guilly H, Dudley RK, Jonard G, Balazs E, Richards KE: Transcription of cauliflower mosaic virus DNA: detection of promoter sequences, and characterization of transcription. Cell 30: 763–773 (1982).
Hernandez G, Cannon F, Cannon M: The effect of presumptive polyadenylation signals on the expression of the CAT gene in transgenic tobacco. Plant Cell Rep 8: 195–198 (1989).
Ingelbrecht ILW, Herman LMF, Dekeyser RA, Van Montagu MC, Depicker AG: Different 3′ end regions strongly influence the level of gene expression in plant cells. Plant Cell 1: 671–680 (1989).
Iturriaga G, Jefferson RA, Bevan MW: Endoplasmic reticulum targeting and glycosylation of hybrid proteins in transgenic tobacco. Plant Cell 1: 381–390 (1989).
Jefferson RA: Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 (1987).
Jefferson RA, Kavanagh TA, Bevan MW: GUS Fusions:β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Jefferson RA, Wilson KJ: The GUS gene fusion system. In: Gelvin SB, Schilperoort RA, Verma DPS (eds). Plant Molecular Biology Manual, pp. B14/1-B14/33. Kluwer Academic Publishers, Dordrecht, The Netherlands (1991).
Kavanagh TA, Jefferson RA, Bevan MW: Targeting a foreign peptide to chloroplasts using fusions to the transit peptide of a chlorophyll a/b protein. Mol Gen Genet 215: 38–45 (1988).
Koziel MG, Beland GL, Bowman C, Carozzi NB, Crenshaw R, Crossland L, Dawson J, Desai N, Hill M, Kadwell S, Launis K, Lewis K, Maddox D, McPherson K, Meghji MR, Merlin E, Rhodes R, Warren GW, Wright M, Evola SV: Field performance of elite transgenic maize plants expressing an insecticidal protein derived fromBacillus thuringiensis. Bio/technology 11: 194–200 (1993).
Kyozuka J, McElroy D, Hayakawa T, Xie Y, Wu R, Shimamoto K: Light- regulated and cell specific expression of tomatorbcS-gus and ricerbcS-gus fusion genes in transgenic rice. Plant Physiol 102: 991–1000 (1993).
Luehrsen KR, Walbot V: Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol Gen Genet 225: 81–93 (1991).
McElroy D, Rothenberg M, Reece KS, Wu R: Characterization of the rice (Oryza sativa) actin gene family. Plant Mol Biol 15: 257–268 (1990).
McElroy D, Zhang W, Cao J, Wu R: Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163–171 (1990).
McElroy D, Blowers AD, Jenes B, Wu R: Construction of expression vectors based on the rice actin 1 (Act1) 5′ region for use in monocot transformation. Mol Gen Genet 231: 150–160 (1991).
McElroy D, Brettell RIS: Foreign gene expression in transgenic cereals: progress and pitfalls. Trends Biotechnol 12: 62–68 (1994).
Mogen BD, MacDonald MH, Graybosch R, Hunt AG: Upstream sequences other than AAUAAA are required for efficient messenger RNA 3′-end formation in plants. Plant Cell 2: 1261–1272 (1990).
Peterhans A, Datta SK, Datta K, Goodall GJ, Potrykus I, Paszkowski J: Recognition efficiency of Dicotyledoneae-specific promoter and RNA processing signals in rice. Mol Gen Genet 222: 361–368 (1991).
Pierce DA, Mettler IJ, Lachmansingh AR, Pomeroy LM, Weck EA, Mascarenhas D: Effects of 35S leader modification on promoter activity. In: McIntosh L, Key J (eds) Plant Gene Systems and Their Biology, pp. 301–310. Alan R. Liss, New York (1987).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Somers DA, Rines HW, Gu W, Kaeppler HF, Bushnell WR: Fertile, transgenic oat plants. Bio/technology 10: 1589–1594 (1992).
Vasil V, Clancy M, Ferl R, Vasil IK, Hannah LC: Increased gene expression by the first intron of maizeshrunken-1 locus in grass species. Plant Physiol 91: 1575–1579 (1989).
Vasil V, Castillo AM, Fromm ME, Vasil IK: Herbicide resistant fertile transgenic wheat plants obtained by microparticle bombardment of regenerable embryogenic callus. Bio/technology 10: 667–574 (1992).
Wan Y, Lemaux PG: Generation of large numbers of independently transformed fertile barley plants. Plant Physiol 104: 37–48 (1994).
Weeks T, Anderson OD, Blechl AE: Rapid production of multiple independent lines of fertile transgenic wheat (Triticum aestivum). Plant Physiol 102: 1077–1084 (1993).
Xie Y, Pen Z, Cai Y, Wu R: Cloning and analysis of histone 3 gene and rubisco small subunit gene of rice. Sci Sinica (ser B) 30: 706–718 (1987).
Xu D, McElroy D, Thornburg R, Wu R: Systemic induction of a potatopin2 promoter by wounding, methyl jasmonate, and abscisic acid in transgenic rice plants. Plant Mol Biol 22: 573–588 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
McElroy, D., Chamberlain, D.A., Moon, E. et al. Development ofgusA reporter gene constructs for cereal transformation: Availability of plant transformation vectors from the CAMBIA Molecular Genetic Resource Service. Mol Breeding 1, 27–37 (1995). https://doi.org/10.1007/BF01682087
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01682087