Abstract
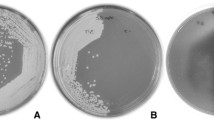
Recombinant Saccharomyces cerevisiae strains capable of simultaneous secretion of bacterial glucanase and pectinase enzymes have been developed. The Butyrivibrio fibrisolvens endo-β-1,4-glucanase gene (end1), the Erwinia chrysanthemi pectate lyase gene (pelE) and E. carotovora polygalacturonase gene (peh1) were each inserted between a yeast expression-secretion cassette and yeast gene terminator, and cloned into yeast-centromeric shuttle vectors. Transcription initiation signals present in the expression-secretion cassette were derived from the yeast alcohol dehydrogenase gene promoter (ADC1 P ), whereas the transcription termination signals were derived from the yeast tryptophan synthase gene terminator (TRP5 T ). Secretion of glucanase and pectinases was directed by the signal sequence of the yeast mating pheromone α-factor (MFα1 S ). These YCplac111-based constructs, designated END1, PEL5, and PEH1, respectively, were transformed into S. cerevisiae. The END1, PEL5 and PEH1 constructs were co-expressed in laboratory strains of S. cerevisiae as well as in wine and distillers' yeasts. DNA-RNA hybridization analysis showed the presence of END1, PEL5 and PEH1 transcripts. Carboxymethylcellulose and polypectate agarose assays revealed the production of biologically active endo-β-1,4-glucanase, pectate lyase and polygalac-turonase by the S. cerevisiae transformants. Interestingly, although the same expression-secretion cassette was used in all three constructs, time-course assays indicated that the pectinases were secreted before the glucanase. It is tempting to speculate that the bulkiness of the END1-encoded protein and the five alternating repeats of Pro-Asp-Pro-Thr(Gln)-Pro-Val-Asp within the glucanase moiety could be involved in the delayed secretion of the glucanase.
Similar content being viewed by others
References
Ammerer G (1983) Methods Enzymol 101:192–210
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1987) Current protocols in molecular biology. John Wiley and Sons, New York
Béguin P, Gilkes NR, Kilburn DG, Miller RC, O'Neil GP, Warren RAJ (1987) Crit Rev Biotechnol 6:129–162
Beier RD, Young ET (1982) Nature 300:724–728
Berger E, Jones WA, Jones DT, Wood DR (1989) Mol Gen Genet 219:193–198
Beldman G, Rombouts FM, Voragen AGJ, Pilnik W (1984) Enzyme Microb Technol 6:503–507
Bell TA, Etchells JL (1956) Appl Microbiol 4:196–202
Bitter GA, Chen KK, Banks AR, Lai P (1984) Proc Natl Acad Sci USA 81:5330–5334
Bitter GA, Egan KM, Koski RA, Jones MO, Elliott SG, Giffin JC (1989) Methods Enzymol 153:516–544
Brake AJ (1990) Methods Enzymol 185:408–421
Brown MR, Ough CS (1981) Am J Vitic Enol 32:272–276
Coughlan MP (1989) Enzyme systems for lignocellulose degradation. Commission of the European Communities. Elsevier Applied Science, London
Cowling EB, Kirk TK (1976) Properties of cellulose and lignocellulosic materials as substrates for enzymatic conversion processes. In: Gaden EL Jr, Mandels MH, Reese ET, Spano LA (eds) Enzymatic conversion of cellulosic materials: technology and applications. John Wiley and Sons, New York, pp 95–123
Denis CL, Ferguson J, Young ET (1983) J Biol Chem 258: 1156–1171
Farkas V, Biely P, Bauer S (1973) Biochim Biophys Acta 321: 246–255
Ghose TK (1987) Pure Appl Chem 59:257–268
Gietz RD, Schiestl RH (1991) Yeast 7:253–263
Gietz RD, Sugino A (1988) Gene 74:527–534
Hitzeman RA, Hagie FE, Levine HL, Goeddel DV, Ammerer G, Hall BD (1981) Nature 293:717–722
Hong JC, Nagao RT, Key JL (1990) J Biol Chem 265:2470–2475
Innis MA, Gelfand DH, (1990) Optimizing of PCRs. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols. Academic Press, San Diego, California, pp 3–12
Janse BJH, Pretorius IS (1993) Curr Genet 24:32–37
Keen NT, Dahlbeck D, Staskawicz B, Belser W (1984) J Bacteriol 159:825–831
Klebl F, Tanner W (1989) J Bacteriol 171:6259–6264
Kuranda MJ, Robbins P (1987) Proc Natl Acad Sci USA 84:2585–2589
Kurjan J, Herskowitz I (1982) Cell 30:933–943
Laing E, Pretorius IS (1992) Gene 121:35–45
Laing E, Pretorius IS (1993a) J Appl Bacteriol 75:149–158
Laing E, Pretorius IS (1993b) Appl MicrobiolBiotechnol 39: 181–188
Luh BS, Phaff HJ (1951) Arch Biochem Biophys 33:212–227
MacMillan JD, Sheiman MI (1980) Pectic enzymes. In: Whitaker JR (ed) Food related enzymes. American Chemical Society, Washington, D.C., pp 101–130
Marcus A, Greenberg J, Averyhart-Fullard V (1991) Physiol Plant 81:273–279
Mrsa V, Klebl F, Tanner W (1993) J Bacteriol 175:2102–2106
Nebreda AR, Villa TG, Villanueva JR, del Rey F (1986) Gene 47:245–259
Pilnik W, Rombouts FM (1979) Pectic enzymes. In: Blanshard JMV, Mitchell JR (eds) Polysaccharides in food. Butterworths, Boston, pp 109–126
Pretorius IS, Van der Westhuizen TJ (1991) S Afr J Enol Vitic 12:3–31
Pretorius IS, Chow T, Modena D, Marmur J (1986) Mol Gen Genet 203:29–35
Pretorius IS, Laing E, Pretorius GHJ, Marmur J (1988) Curr Genet 14:1–8
Rankine B (1991) The Aust Graper Winemaker 334:13
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA (1988) Science 239:287–491
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd. edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Saulnier L, Brillouet J-M (1988) Carbohydr Res 182:63–78
Southgate VJ, Steyn AJC, Pretorius IS, Van Vuuren HJJ (1993) Appl Environ Microbiol 59:1253–1258
Steyn AJC, Pretorius IS (1991) Gene 100:85–93
Teather RM, Wood PJ (1982) Appl Environ Microbiol 53:41–46
Willis JW, Engwall JK, Chatterjee AK (1987) Phytopathology 77:1199–1205
Wood TM, Garcia-Campayo V (1990) Biodegradation 1:147–161
Wright RM, Yablonsky MD, Shalita ZP, Goyal AK, Eveleigh DE (1992) Appl Environ Microbiol 58:3455–3465
Yablonsky MD, Bartley T, Elliston KO, Kahrs SK, Shalita ZP, Eveleigh DE (1988) FEMS Sym 43:249–266
Author information
Authors and Affiliations
Additional information
Communicated by B. S. Cox
Rights and permissions
About this article
Cite this article
van Rensburg, P., van Zyl, W.H. & Pretorius, I.S. Expression of the Butyrivibrio fibrisolvens endo-β-1,4-glucanase gene together with the Erwinia pectate lyase and polygalacturonase genes in Saccharomyces cerevisiae . Curr Genet 27, 17–22 (1994). https://doi.org/10.1007/BF00326573
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00326573