Abstract
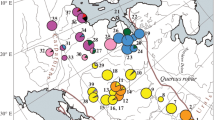
Chloroplast DNA polymorphisms have been detected by the conventional Southern-blotting hybridization method in four species of European oaks (Quercus petraea, Q. robur, Q. pubescens and Q. pyrenaica). Three polymorphisms, shared by at least three of these species, can be scored directly in ethidium bromidestained gels and were used in a broad survey of the level of differentiation of the oak species and of their pattern of genetic structure in western Europe. The highly significant geographic variation and the high genetic differentiation (Gst=0.895, SGst=0.025) indicate a low level of cytoplasmic gene flow. We conclude that cytoplasmic genomes are well suited for the reconstruction of past migrational routes of such a complex of species.
Similar content being viewed by others
References
Bossema I (1979) Jays and oaks: an eco-ethological study of a symbiosis. Behaviour 70:1–117
Chakraborty R, Leimar O (1987) Genetic variation within a subdivided population. In: Ryman N, Utter F (eds) Population genetics and fishery management. Washington Sea Grant program, Seattle and London, pp 89–120
Chiu W-L, Sears BB (1985) Recombination between chloroplast DNAs does not occur in sexual crosses of Oenothera. Mol Gen Genet 198:525–528
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Efron B (1979) Bootstrap methods: another look at the jackknife. Ann Stat 7:1–26
Engels WR (1981) Estimating genetic divergence and genetic variability with restriction endonucleases. Proc Natl Acad Sci USA 78:6329–6333
Hamrick JL, Godt MJW (1990) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetic resources. Sinauer associates Inc., Sunderland, Massachusetts, pp 43–63
Huntley B, Birks HJB (1983) An atlas of past and present pollen maps for Europe, 0–13,000 years ago. Cambridge University Press, Cambridge, UK
Kremer A, Petit RJ, Zanetto A, Fougère V, Ducousso A, Wagner D (1991) Nuclear and organelle gene diversity in Q. robur and Q. petraea. In: Ziehe M, Müller-Starck G (eds) Genetic variation of forest tree populations in Europe. Sauerländer's Verlag, Frankfurt am Main, pp 141–172
Moran PAP (1950) Notes on continuous stochastic phenomena. Biometrika 37:17–23
Müller-Starck G, Herzog S, Hattemer HH (1993) Intraand interpopulational genetic variation in juvenile populations of Quercus robur L. and Quercus petraea Liebl. Ann Sci For 50 (Suppl 1):233s-244s
Nei M (1977) F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet 41:225–233
Nei M (1987) Molecular evolutionary genetics. Colombia University Press, New York
Ohri D, Ahuja MR (1990) Giemsa C-banded karyotype in Quercus L. (oak). Silvae Genet 39:216–219
Palmer JD (1986) Isolation and structural analysis of chloroplast DNA. Methods Enzymol 118:167–186
Palmer JD, Shields CR, Cohen DB, Orton TJ (1983) Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor Appl Genet 65:181–189
Šiďak Z (1967) Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc 62:626–633
Smith JF, Doyle JJ (1986) Chloroplast DNA variation and evolution in the Juglandaceae. Am J Bot 73:730
Sokal RR, Oden NL (1978) Spatial auto-correlation in biology. 1. Methodology. Biol Jour Linnean Soc 10:199–228
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Sytsma KJ, Schaal BA (1985) Phylogenetics of the Lisianthus skinneri (Gentianaceae) species complex in Panama utilizing DNA restriction fragment analysis. Evolution 39:594–608
Wagner DB, Sun Z-X, Govindaraju DR, Dancik BP (1991) Spatial patterns of chloroplast DNA and cone morphology variation within populations of a Pinus banksiana-Pinus contorta sympatric region. Am Nat 138:156–170
Wilson MA, Gaut B, Clegg MT (1990) Chloroplast DNA evolves slowly in the palm family. Mol Biol Evol 7:303–314
Author information
Authors and Affiliations
Additional information
Communicated by P. M. A. Tigerstedt
Rights and permissions
About this article
Cite this article
Petit, R.J., Kremer, A. & Wagner, D.B. Geographic structure of chloroplast DNA polymorphisms in European oaks. Theoret. Appl. Genetics 87, 122–128 (1993). https://doi.org/10.1007/BF00223755
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00223755