Abstract
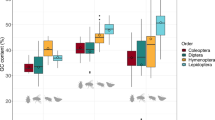
The abundance and polymorphism of 38 different simple-sequence repeat motifs was studied in four accessions of cultivated chickpea (Cicer arietinum L.) by in-gel hybridization of synthetic oligonucleotides to genomic DNA digested with 14 different restriction enzymes. Among 38 probes tested, 35 yielded detectable hybridization signals. The abundance and level of polymorphism of the target sequences varied considerably. The probes fell into three broad categories: (1) probes yielding distinct, polymorphic banding patterns; (2) probes yielding distinct, monomorphic banding patterns, and (3) probes yielding blurred patterns, or diffused bands superimposed on a high in lane background. No obvious correlation existed between abundance, fingerprint quality, and the sequence characteristics of a particular motif. Digestion with methyl-sensitive enzymes revealed that simple-sequence motifs are enriched in highly methylated genomic regions. The high level of intraspecific polymorphism detected by oligonucleotide fingerprinting suggests the suitability of simple-sequence repeat probes as molecular markers for genome mapping.
Similar content being viewed by others
References
Ali S, Müller CR, Epplen JT (1986) DNA fingerprinting by oligonucleotide probes specific for simple repeats. Hum Genet 74:239–243
Broun P, Tanksley SD (1993) Characterization of tomato DNA clones with sequence similarity to human minisatellites 33.6 and 33.15. Plant Mol Biol 23:231–242
Epplen JT (1988) On simple repeated GATA/GACA sequences in animal genomes: a critical reappraisal. J Hered 79:409–417
Gaur PM, Slinkard AE (1990a) Inheritance and linkage of isozymecoding genes in chickpea. J Hered 81:455–461
Gaur PM, Slinkard AE (1990b) Genetic control and linkage relations of additional isozyme markers in chickpea. Theor Appl Genet 80:648–656
Georges M, Lathrop M, Hilbert P, Marcotte A, Schwers A, Swillens S, Vassart G, Hanset R (1990) On the use of DNA fingerprints for linkage studies in cattle. Genomics 6:461–474
Jeffreys AJ, Royle NJ, Wilson V, Wong Z (1988) Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332:278–281
Julier C, De Gouyon B, Guénet M, Guénet JL, Nakamura Y, Avner P, Lathrop GM (1990) Minisatellite linkage maps in the mouse by cross-hybridization with human probes containing tandem repeats. Proc Natl Acad Sci USA 86:4585–4589
Kazan K, Muehlbauer FJ (1991) Allozyme variation and phytogeny in annual species of Cicer (Leguminosae). Plant Syst Evol 175:11–21
Kazan K, Muehlbauer FJ, Weeden NF, Ladizinsky G (1993) Inheritance and linkage relationships of morphological and isozyme loci in chickpea (Cicer arietinum L.). Theor Appl Genet 86:417–426
Lagercrantz U, Ellegren H, Andersson L (1993) The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res 21:1111–1115
Melchinger AE (1990) Use of molecular markers in breeding for oligogenic disease resistance. Plant Breed 104:1–19
Nanda I, Deubelbeiss C, Guttenbach M, Epplen JT, Schmid M (1990) Heterogeneities in the distribution of (GACA) n simple repeats in the karyotypes of primates and mouse. Hum Genet 85:187–194
Romao J, Hamer JE (1992) Genetic organization of a repeated DNA sequence family in the rice blast fungus. Proc Natl Acad Sci USA 89:5316–5320
Rychlik W, Rhoads RE (1989) A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in-vitro amplification of DNA. Nucleic Acids Res 17:8543–8551
Saxena MC, Singh KB (1987) The chickpea. CAB International, Wallingford, UK
Singh KB, Saxena MC (1992) Disease resistance in chickpea. ICARDA, Aleppo, Syria
Singh KB, Reddy MV, Haware MP (1992) Breeding for resistance to Ascochyta blight in chickpea. In: Singh KB, Saxena MC (eds) Disease resistance in chickpea. ICARDA, Aleppo, Syria, pp 23–54
Thein SL, Wallace RB (1986) The use of synthetic oligonucleotides as specific hybridization probes in the diagnosis of genetic disorders. In: Davies KE (ed) Human genetic diseases-a practical approach. IRL Press, Oxford, pp 33–50
Weising K, Weigand F, Driesel A, Kahl G, Zischler H, Epplen JT (1989) Polymorphic simple GATA/GACA repeats in plant genomes. Nucleic Acids Res 17:10128
Weising L, Beyermann B, Ramser J, Kahl G (1991) Plant DNA finger-printing with radioactive and digoxigenated oligonucleotide probes complementary to simple repetitive sequences. Electrophoresis 12:159–169
Weising K, Kaemmer D, Weigand F, Epplen JT, Kahl G (1992) Oligonucleotide fingerprinting reveals various probe-dependent levels of informativeness in chickpea (Cicer arietinum). Genome 35:436–442
Wells RA, Green P, Reeders ST (1989) Simultaneous mapping of multiple human minisatellite sequences using DNA fingerprinting Genomics 5:761–772
Willmitzer L, Wagner KG (1981) The isolation of nuclei from tissue-cultured plant cells. Exp Cell Res 135:69–77
Author information
Authors and Affiliations
Additional information
Communicated by G. Wenzel
Rights and permissions
About this article
Cite this article
Sharma, P.C., Winter, P., Bünger, T. et al. Abundance and polymorphism of di-, tri-and tetra-nucleotide tandem repeats in chickpea (Cicer arietinum L.). Theoret. Appl. Genetics 90, 90–96 (1995). https://doi.org/10.1007/BF00221000
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00221000