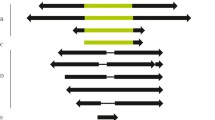
Summary
Characterization of sequences homologous to theDrosophila melanogaster gypsy transposable element was carried out inDrosophila subobscura (gypsyDS). They were found to be widely distributed among natural populations of this species. From Southern blot and in situ analyses, these sequences appear to be mobile in this species.GypsyDS sequences are located in both euchromatic and heterochromatic regions. A completegypsyDS sequence was isolated from aD. subobscura genomic library, and a 1.3-kb fragment which aligns with the ORF2 of theD. melanogaster gypsy element was sequenced. Comparisons of this sequence in three species (D. subobscura, D. melanogaster, and D. virilis) indicate that there is greater similarity between theD. subobscura-D. virilis sequences than betweenD. subobscura andD. melanogaster. Molecular divergence ofgypsy sequences betweenD. virilis andD. subobscura is estimated at 16 MY, whereas the most likely divergence time of these two species is more than 60 MY. These data strongly suggest thatgypsy sequences have been horizontally transferred between these species.
Similar content being viewed by others
References
Abad P, Vaury C, Pelisson A, Chaboissier MC, Busseau I, Bucheton A (1989) A long interspersed repetitive element—theI factor ofDrosophila teissieri—is able to transpose in differentDrosophila species. Proc Natl Acad Sci USA 86: 8887–8891
Anxolabéhère D, Kidwell MG, Periquet G (1988) Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion ofDrosophila melanogaster by mobileP elements. Mol Biol Evol 5:252–269
Bajev AA, Lyubomirskaya NV, Dzhumagaliev EB, Ananiev EV, Amiantova IG, Ilyin YV (1984) Structural organization of transposable elementmdg4 fromDrosophila melanogaster and a nucleotide sequence of its long terminal repeats. Nucl Acid Res 12:3707–3723
Beckenbach AT, Prevosti A (1986) Colonization of North America by the European species,Drosophila subobscura andDrosophila ambigua. Am Midland Naturalist 115:10–18
Bingham PM, Zachar Z (1989) Retrotransposons and theFB transposon fromDrosophila melanogaster. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington, DC, pp 485–502
Blackman RK, Meselson M (1986) Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of theDrosophila hsp82 gene. J Mol Biol 188: 499–515
Brncic D, Prevosti A, Budnik M, Ocaña J (1981) Colonization ofDrosophila subobscura in Chile I. First population and cytogenetic studies. Genetica 56:3–9
Brookfield JFY, Montgomery E, Langley CH (1984) Apparent absence of transposable elements related to theP elements ofDrosophila melanogaster in other species ofDrosophila. Nature 310:330–332
Csink AK, McDonald JF (1990)Copia expression is variable among natural populations ofDrosophila. Genetics 126:375–385
Daniels SB, Strausbaugh LD (1986) The distribution ofP element sequences inDrosophila: thewillistoni andsaltans species groups. J Mol Evol 23:138–148
Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A (1990a) Evidence for horizontal transmission of theP transposable element betweenDrosophila species. Genetics 124:339–355
Daniels SB, Chovnick A, Boussy IA (1990b) Distribution ofhobo transposable elements in the genus Drosophila. Mol Biol Evol 7:589–606
Doolittle RF, Feng DF, Johnson MS, McClure MA (1989) Origins and evolutionary relationships of retroviruses. Quart Rev Biol 64:1–30
Engels WR (1989)P elements inDrosophila melanogaster. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington, DC, pp 437–484
Finnegan D, Fawcett D (1986) Transposable elements inDrosophila melanogaster. In: Maclean N (ed) Oxford surveys on eukaryotic genes, vol 3, pp 1–62
Finnegan DJ (1989) The I factor and I-R hybrid dysgenesis inDrosophila melanogaster. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington, DC, pp 503–517
de Frutos R, Peterson KR, Kidwell MG (1992) Distribution ofDrosophila melanogaster transposable element sequences in species of theobscura group. Chromosoma 101:293–300
Hartl DL, Clark AG (1988) Principles of population genetics. Sinauer, Sunderland, MA, pp 682
Kidwell MG, Kidwell JF, Sved JA (1977) Hybrid dysgenesis inDrosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86:813–833
Kidwell MG (1979) Hybrid dysgenesis inDrosophila melanogaster: the relationship between the P-M and I-R interaction systems. Genet Res 33:105–117
Kidwell MG (1983) Evolution of hybrid dysgenesis determinants inDrosophila melanogaster. Proc Natl Acad Sci USA 80: 1655–1659
Kidwell MG (1992) Horizontal transfer of P elements and other short inverted repeat transposons. Genetica (in press)
Lansman RA, Stacey SN, Grigliatti TA, Brock HW (1985) Sequences homologous to theP mobile element ofDrosophila melanogaster are widely distributed in the subgenus Sophophora. Nature 318:561–563
McDonald JF (1989) The potential evolutionary significance of retroviral-like transposable elements in peripheral populations. In: Fontdevila A (ed) Evolutionary biology of transient unstable populations. Springer-Verlag, New York, pp 190–205
Martin G, Wiernasz D, Schedl P (1983) Evolution ofDrosophila repetitive-dispersed DNA. J Mol Evol 19:203–213
Marlor RL, Parkhurst SM, Corces VG (1986) TheDrosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol 6:1129–1134
Mizrokhi LJ, Mazo AM (1990) Evidence for horizontal transmission of the mobile elementjockey between distantDrosophila species. Proc Natl Acad Sci USA 87:9216–9220
Mizrokhi LJ, Mazo AM (1991) Cloning and analysis of the mobile elementgypsy andDrosophila virilis. Nucl Acid Res 19: 913–916
Modolell J, Bender W, Meselson M (1983)Drosophila melanogaster mutations suppressible by the suppressor ofHairywing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci USA 80:1678–1682
Paricio N, Perez-Alonso M, Martinez-Sebastian MJ, de Frutos R (1991)P sequences ofDrosophila subobscura lack exon 3 and may encode a 66 kd repressor-like protein. Nucl Acid Res 19:6713–6718
Pascual L, Periquet G (1991) Distribution ofhobo transposable elements in natural populations ofDrosophila melanogaster. Mol Biol Evol 8:282–296
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci 74:5463–5467
Silber J, Bazin C, Lemeunier F, Aulard S, Volovitch M (1989) Distribution and conservation of the Foldback transposable element inDrosophila. J Mol Evol 28:220–224
Seeger MA, Kaufman TC (1990) Molecular analysis of thebicoid gene fromD. pseudoobscura: identification of conserved domains within coding and noncoding regions of thebicoid mRNA. EMBO J 9:2977–2987
Stacey SN, Lansman RA, Brock HW, Grigliatti TA (1986) Distribution and conservation of mobile elements in the genusDrosophila. Mol Biol Evol 3:522–534
Tabor S, Richardson CC (1987) DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA 84:4767–4771
Terol J, Perez-Alonso M, de Frutos R (1991) In situ localization of theAntennapedia gene on the chromosomes of nineDrosophila species of theobscura group. Hereditas 114:131–139
Throckmorton LH (1975) The phylogeny, ecology and geography ofDrosophila. In: King RC (ed) Handbook of genetics, vol 3. Plenum Press, New York, pp 421–469
Wilde CD, Akam M (1987) Conserved sequence elements in the 5′ region of theUltrabithorax transcription unit. EMBO J 6:1393–1401
Author information
Authors and Affiliations
Additional information
Offprint requests to: T.M. Alberola
Rights and permissions
About this article
Cite this article
Alberola, T.M., de Frutos, R. Gypsy homologous sequences inDrosophila subobscura (gypsyDS). J Mol Evol 36, 127–135 (1993). https://doi.org/10.1007/BF00166248
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00166248