Abstract
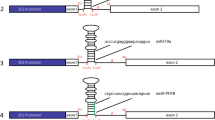
When exogenous genes are to be expressed in transgenic plants, their RNAs must be correctly processed. To gain information useful for predicting whether foreign introns will be accurately spliced, we have analysed the processing of an artificial gene in maize and Nicotiana plumbaginifolia protoplasts. A synthetic plant intron, devised to contain the elements necessary for pre-mRNA splicing in dicots, was found to be efficiently spliced in a monocot (maize) transient expression system. A series of deletion mutants of the synthetic intron was constructed to assess the minimum functional intron length. In both monocots and dicots this was found to be between 70 and 73 nt. This length requirement is similar to that seen in vertebrates, but significantly greater than that in fungi and insects.
Similar content being viewed by others
References
Green MR: Pre-mRNA splicing. Ann Rev Genet 20: 671–708 (1986).
Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Ann Rev Biochem 55: 1119–1150 (1986).
Langford CJ, Klinz F-J, Donath C, Gallwitz D: Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell 36: 645–653 (1984).
Frendewey D, Keller W: The stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 42: 355–367 (1985).
Ruskin B, Green MR: Role of the 3′ splice site consensus sequence in mammalian pre-mRNA splicing. Nature 317: 732–734 (1985).
Goodall GJ, Filipowicz W: The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483 (1989).
Hawkins JD: A survey on intron and exon lengths. Nucl Acids Res 16: 9893–9908 (1988).
Martinez-Zapater JM, Finkelstein R, Somerville CR: Drosophila P-element transcripts are incorrectly processed in tobacco. Plant Mol Biol 11: 601–607 (1988).
Laski FA, Rio DC, Rubin GM: Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell 44: 7–19 (1986).
Goodall GJ, Wiebauer K, Filipowicz W: Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol 181, in press (1989).
Keith B, Chua N-H: Monocot and dicot pre-mRNAs are processed with different efficiencies in transgenic tobacco. EMBO J 5: 2419–2425 (1986).
Wieringa B, Hofer E, Weissmann C: A minimal intron length but no specific internal sequence is required for splicing the large rabbit β-globin intron. Cell 37: 915–925 (1984).
Gallwitz D, Halfter H, Mertins P: Splicing of mRNA precursors in yeast. In: Kinghorn JA (ed) Gene Structure in Eukaryotic Microbes, pp. 27–40. IRL Press, Oxford/Washington DC (1987).
Ruskin B, Greene JM, Green MR: Cryptic branch point activation allows accurate in vitro splicing of human β-globin intron mutants. Cell 41: 833–844 (1985).
Steitz JA, Black DL, Gerke V, Parker KA, Krämer A, Frendewey D, Keller W: Functions of the abundant U-snRNPs. In: Birnstiel ML (ed) Small Nuclear Ribonucleoprotein Particles, pp. 115–154. Springer-Verlag, Berlin (1988).
Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H: Molecular analysis of the En/Spm transposable element system of Zea mays. EMBO J 5: 835–841 (1986).
Takaei Y, Yamauchi D, Minamikawa T: Nucleotide sequence of the canavalin gene from Canavalia gladiata seeds. Nucl Acids Res 17: 4381 (1989).
Walbot V, Britt AB, Luehrsen K, McLaughlin M, Warren C: Regulation of mutator activities in maize. In: Oliver Nelson (ed) Plant Transposable Elements, pp. 121–135. Plenum Press, New York (1988).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Goodall, G.J., Filipowicz, W. The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol Biol 14, 727–733 (1990). https://doi.org/10.1007/BF00016505
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00016505