Abstract
We propose a new regularization for spherical deconvolution in diffusion MRI. It is based on observing that higher-order tensor representations of fiber ODFs should be H-psd, i.e., they should have a positive semidefinite (psd) matrix \(H_T\). We show that this constraint is stricter than the currently more widely used non-negativity, and that it can be enforced easily using quadratic cone programming. We demonstrate its use in a multi-tissue deconvolution framework that models the different tissue types in the continuous SHORE basis and can therefore be applied to data with multiple b values that are not organized on shells, such as in Diffusion Spectrum Imaging. Experiments on simulated fiber crossings, data from the Human Connectome Project, and clinical data, demonstrate the improved speed and accuracy of this new method.
S. Groeschel—This work was supported by the DFG under grant SCHU 3040/1-1. LHL is supported by AFOSR FA9550-13-1-0133, DARPA D15AP00109, NSF IIS 1546413, DMS 1209136, and DMS 1057064. Parts of the data (“clin-dsi”) were provided by Katrin Sakreida and Georg Neuloh (RWTH Aachen University Hospital), and by the HCP, WU-Minn Cons. (PIs: D. Van Essen and K. Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington U.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Spherical deconvolution [14] is widely used to analyze white matter (WM) in high angular resolution diffusion imaging (HARDI). Recently, it has been shown that, when HARDI data is available on multiple shells, parameters related to the volume fractions of gray matter (GM) and corticospinal fluid (CSF) can be added to the deconvolution model [3, 5]. An important advantage of this is increased accuracy in voxels with partial voluming between different tissue types.
We present a novel method for multi-tissue deconvolution that makes two main contributions: First, non-negativity constraints have long been recognized as an indispensible regularization in deconvolution, and are most frequently enforced at a set of discrete points on the sphere [14, 16]. In contrast, we derive a positive definiteness constraint from a higher-order tensor representation of the fiber orientation distribution function (fODF) which implies non-negativity everywhere on the sphere. It can be implemented easily using quadratic cone programming, and can be combined both with multi-tissue and traditional single-shell single-tissue deconvolution.
Second, many dMRI acquisition schemes do not use a shell structure. This includes Diffusion Spectrum Imaging [15], which uses a Cartesian grid, and some recently proposed schemes that distribute samples freely in Q-space [7, 9]. Our approach improves upon previous multi-tissue deconvolution methods, which have modeled the signal on each shell separately [3, 5], by instead estimating the tissue response as a continuous function in Q-space, using the SHORE basis functions [8]. This allows us to work with data that is not organized on shells.
In a direct comparison between our approach and a state-of-the-art alternative [5], we demonstrate that ours is more well-conditioned, more accurate at small angles, and faster.
2 Related Work
Our approach extends previous work that has used higher-order Cartesian tensors to represent fiber ODFs [6, 11, 13, 16] by the ability to handle data acquired at multiple b-values and by modeling multiple tissue types.
Our H-psd constraint is new, and stronger than the non-negativity constraints used previously when estimating higher-order diffusion tensors [1, 4], or in the non-negative spherical deconvolution (NNSD) [2] approach. In addition, in contrast to NNSD, our method can be implemented using standard convex cone optimization packages rather than requiring a custom implementation of a computationally expensive Riemannian gradient descent.
3 Fiber ODFs are H-psd Tensors
Our approach extends previous work [13], which has proposed to describe fODFs f by fully symmetric fourth order tensors:
Such fODF tensors T are obtained by deconvolution in the Spherical Harmonics basis and a subsequent change to the monomial basis, which spans the same space of functions. However, there are two important differences to standard spherical deconvolution [14]: First, the deconvolution step is constructed so that it maps the single fiber response in direction \({\mathbf {v}}\) to a symmetric rank-one tensor \({\mathbf {v}}\otimes {\mathbf {v}}\otimes {\mathbf {v}}\otimes {\mathbf {v}}\), rather than to a truncated delta peak. Second, principal fODF directions for fiber tractography are found using a low-rank approximation of the fODF tensor, rather than peaks in the fODF function.
The set of symmetric three-dimensional fourth-order tensors will be denoted by \(S_{3,4}\). The subset of positive semidefinite tensors is
As pointed out in Chap. 5 of [10], \(T \in S_{3,4}\) has an associated matrix
that represents T’s action on the six-dimensional space of symmetric products \({\mathbf {v}} \otimes {\mathbf {v}}\). We will denote the subset of tensors with a positive semidefinite matrix \(H_T\) as
and call these tensors H-psd.
\(P_H\) is closely related to decomposable tensors. To show this, let
be the subsets of tensors that are sums of fourth powers and sums of squares, respectively. Due to their geometric structure, these sets are called cones. They obey
Obviously, sums of squares and sums of fourth powers are positive semidefinite. In fact, a result by Hilbert, which has been applied to enforce non-negativity of fourth-order diffusion tensors [1, 4], states that \(P_{3,4} = \varSigma _{3,4}\). In contrast, not every tensor in \(P_{3,4}\) can be written as a sum of fourth powers [10].
The dual of a cone C is defined as \( C^\star = \{ y \in C : \langle x,y \rangle \ge 0 \; \forall x \in C \} \) and, in [10], it is shown that
Combining these dualities with the dual of the result by Hilbert yields
i.e., a tensor in \(S_{3,4}\) can be written as a sum of fourth powers or, equivalently, decomposed into symmetric rank-1 terms, if and only if it is H-psd.
For spherical deconvolution, this has the following implications: First, fODF tensors arise from a mixture of an unknown number of single-fiber compartments, each of which is represented as a symmetric rank-1 tensor. Therefore, any valid fODF tensor must be decomposable. Second, decomposability can be checked easily by positive semidefiniteness of Eq. (3) (H-psd). Third, as Eq. (7) shows, H-psd is a stronger condition than simple non-negativity, as it has been imposed previously on fODFs [2, 14], or on fourth-order diffusion tensors [1, 4].
4 Using SHORE for Multi-tissue Deconvolution
Multi-tissue deconvolution requires modelling the dMRI response of all tissue types. Previous approaches [3, 5] do so separately for each shell. To avoid dependence on a shell structure, we instead create a continuous model of functions \(f(\mathbf{q } = q \mathbf{u })\) in Q-space using the SHORE basis functions [8]
with the associated Laguerre polynomials \(L^\alpha _n\), the real Spherical Harmonics \(Y_l^m\) and a radial scaling factor \(\zeta \).
Let \(K(\mathbf{q }) = \sum _{ln} K_{ln} \, \phi _{ln0}(\mathbf{q })\) be the white matter single-fiber response, with \(m=0\) due to cylinder symmetry. The signal from an fODF f is then modeled by a convolution on the sphere, \(S(\mathbf{q }) \approx K \star _{{\mathbb {S}}^2} f\) [2]. For a given K and signal vector \(S_i = S(\mathbf{q }_i)\), finding the Spherical Harmonics coefficients f via deconvolution then becomes a linear least squares problem with a nonlinear constraint:
with convolution matrix
As in [13], the \(\alpha _l\) are the Spherical Harmonics coefficients of the unit rank-1 tensor along the z-axis.
The problem is equivalent to the quadratic cone program (QCP)
with \(P = M^T M\), \(q = -M^T S\), and a matrix G that first maps f from Spherical Harmonics to the monomial basis, and then to its \(H_T\) matrix, \(G f = H_f\). This QCP can be solved efficiently using cvxopt (cvxopt.org).
Multi-tissue support is added by concatenating individual tissue matrices
Since CSF and GM are isotropic, \(M_{\mathrm {CSF}}\) and \(M_{\mathrm {GM}}\) are single-column matrices. In the QCP, non-negativity constraints are enforced for \(f_{\mathrm {GM}}\) and \(f_{\mathrm {CSF}}\).
5 Results
5.1 Condition and Computational Efficiency
We compared our method to the approach of Jeurissen et al. [5], using cvxopt in both cases. We used data provided by the Human Connectome Project (HCP) with 270 DWIs, 90 each at \(b\approx \{1000,2000,3000\}\) s/mm\(^2\), and a two-shell clinical data set (“clin-2-sh”) with 94 DWIs, 30 at \(b=700\) and 64 at \(b=2000\). We also show results on another clinical data set (“clin-dsi”) with 128 DWIs, acquired on a Cartesian grid with \(b_{{\mathrm {max}}}=3000\). The approach in [5] cannot be applied to this data, since it includes many different b values with few directions each.
Tissue response functions were estimated as described previously [5], based on tissue segmentation of a coregistered \(T_1\) image, and FA thresholds (\({\mathrm {FA}} > 0.7\) for white matter, \({\mathrm {FA}} < 0.2\) for CSF), except that we model the responses in the SHORE basis as described in Sect. 4.
The processing time (on a single CPU core) as well as the condition number of the matrix P in the quadratic program are listed in Table 1. The fact that we use fourth-order fODFs makes the problem much better conditioned. Our method is also significantly faster, despite the fact that we guarantee non-negativity everywhere, instead of enforcing it only at discrete points. We will now demonstrate that, when combined with tensor approximation [13], order 4 is sufficient to resolve crossings, and even more accurate at smaller angles.
5.2 Accuracy on Simulated Data
Evaluating the accuracy of our method requires data for which volume fractions and orientations of crossing fiber compartments are known. From an HCP data set, we extracted the signal from voxels believed to contain a single fiber of white matter, i.e., being within the white matter mask and having \({\mathrm {FA}}>0.7\). The fiber direction for each voxel was estimated by fitting the diffusion tensor model and finding the eigenvector to the highest eigenvalue. Every voxel of simulated data was created by randomly choosing several single fiber voxels and a voxel with \({\mathrm {FA}}<0.2\) from either the grey matter or the CSF mask, and adding their signals with random volume fractions.
For comparison, we evaluated the original method by Jeurissen et al. [5], our proposed method, as well as a hybrid, which uses our novel regularization and tensor decomposition, but models the response functions using Spherical Harmonics for each shell, rather than SHORE.
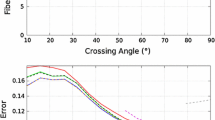
The accuracy of estimated fiber directions and volume fractions on simulated data containing two fibers can be seen in Fig. 1. They demonstrate that, for crossing angles below \(60^\circ \), order-4 tensor approximation reconstructs the fibers much more accurately than finding peaks in order-8 fODFs. The theoretical advantage of H-psd being stricter than non-negativity becomes relevant for small angles, starting around \(40^\circ \). Our simulated data includes the noise from the HCP data from which it was generated. We tried adding more noise, and observed that this increased the advantage of using the H-psd constraint.
Practically no difference is visible when replacing SHORE with Spherical Harmonics models on each shell. This demonstrates that, while SHORE is more flexible in that it does not assume a multi-shell structure, it does not decrease accuracy when data is given on shells.
5.3 Results on Real Data
On the two-shell clinical data, our method and the state-of-the-art approach by Jeurissen et al. [5] produced similar results. The estimated directions of up to three fibers with volume fractions above 0.1 are shown on a map of white matter volume fractions in Fig. 2(a/b), and exhibit little difference.
Averaged over the brain mask, the absolute deviation between tissue volume fractions estimated by the two methods was only 0.005 (CSF), 0.022 (GM), and 0.027 (WM). Averaged over the white matter, the angular deviation of fiber directions, weighted by their volume fractions, was \(7.98^\circ \), which is comparable to our simulation-based estimate of accuracy.
Since the true fiber directions are unknown in this case, we cannot be sure which method is more accurate in practice. However, a clear practical benefit of our method is its increased speed, and the fact that it can also be applied to data that has multiple b values, but no shell structure, as the DSI clinical data, for which results are shown in Fig. 2(c).
6 Conclusion
We introduced H-psd, a new positive definiteness constraint for spherical deconvolution, and showed that it is more stringent than usual non-negativity. It can be combined with all deconvolution methods, multi-tissue as well as the more widely used single-shell single-tissue variant [14]. We showed that the H-psd constraint increases accuracy at small angles, and is easy to enforce using convex cone programming libraries.
We also demonstrated that SHORE can be used as an extension of multi-tissue deconvolution [5]. This allowed us to apply multi-tissue deconvolution to DSI data, without sacrificing accuracy when working with data on shells.
Our method relies on the use of fourth-order tensors to describe fODFs. However, it is clear from the results (and was observed previously in [13]) that fourth-order is enough even for low angles and three-way crossings if we combine it with low-rank tensor approximation.
In the future, we will apply the H-psd constraint in the context of adaptive deconvolution [12] to improve per-voxel estimations of fiber response in patients with demyelinating disease. We expect that this will result in more accurate maps of tissue properties, and more reliable tractography.
References
Barmpoutis, A., Jian, B., Vemuri, B.C., Shepherd, T.M.: Symmetric positive 4th order tensors & their estimation from diffusion weighted MRI. In: Karssemeijer, N., Lelieveldt, B. (eds.) IPMI 2007. LNCS, vol. 4584, pp. 308–319. Springer, Heidelberg (2007)
Cheng, J., Deriche, R., Jiang, T., Shen, D., Yap, P.T.: Non-negative spherical deconvolution (NNSD) for estimation of fiber orientation distribution function in single-/multi-shell diffusion MRI. NeuroImage 101, 750–764 (2014)
Christiaens, D., Maes, F., Sunaert, S., Suetens, P.: Convex non-negative spherical factorization of multi-shell diffusion-weighted images. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 166–173. Springer, Heidelberg (2015). doi:10.1007/978-3-319-24553-9_21
Ghosh, A., Deriche, R., Moakher, M.: Ternary quartic approach for positive 4th order diffusion tensors revisited. In: Proceedings of IEEE International Symposium on Biomedical Imaging, pp. 618–621 (2009)
Jeurissen, B., Tournier, J.D., Dhollander, T., Connelly, A., Sijbers, J.: Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage 103, 411–426 (2014)
Jiao, F., Gur, Y., Johnson, C.R., Joshi, S.: Detection of crossing white matter fibers with high-order tensors and rank-k decompositions. In: Székely, G., Hahn, H.K. (eds.) IPMI 2011. LNCS, vol. 6801, pp. 538–549. Springer, Heidelberg (2011)
Knutsson, H., Westin, C.-F.: Tensor metrics and charged containers for 3D Q-space sample distribution. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013, Part I. LNCS, vol. 8149, pp. 679–686. Springer, Heidelberg (2013)
Merlet, S.L., Deriche, R.: Continuous diffusion signal, EAP and ODF estimation via compressive sensing in diffusion MRI. Med. Image Anal. 17, 556–572 (2013)
Paquette, M., Merlet, S., Gilbert, G., Deriche, R., Descoteaux, M.: Comparison of sampling strategies and sparsifying transforms to improve compressed sensing diffusion spectrum imaging. Magn. Reson. Med. 73(1), 401–416 (2015)
Reznick, B.: Sums of Even Powers of Real Linear Forms. American Mathematical Society, Providence (1992)
Schultz, T., Fuster, A., Ghosh, A., Deriche, R., Florack, L., Lim, L.H.: Higher-order tensors in diffusion imaging. In: Westin, C.F., Vilanova, A., Burgeth, B. (eds.) Visualization and Processing of Tensors and Higher Order Descriptors for Multi-Valued Data. Mathematics and Visualization, pp. 129–161. Springer, Heidelberg (2014)
Schultz, T., Groeschel, S.: Auto-calibrating spherical deconvolution based on ODF sparsity. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013, Part I. LNCS, vol. 8149, pp. 663–670. Springer, Heidelberg (2013)
Schultz, T., Seidel, H.P.: Estimating crossing fibers: a tensor decomposition approach. IEEE Trans. Vis. Comput. Graph. 14(6), 1635–1642 (2008)
Tournier, J.D., Calamante, F., Connelly, A.: Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. NeuroImage 35, 1459–1472 (2007)
Wedeen, V.J., Hagmann, P., Tseng, W.Y.I., Reese, T.G., Weisskoff, R.M.: Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 54(6), 1377–1386 (2005)
Weldeselassie, Y.T., Barmpoutis, A., Atkins, M.S.: Symmetric positive semi-definite cartesian tensor fiber orientation distributions (CT-FOD). Med. Image Anal. 16(6), 1121–1129 (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Ankele, M., Lim, LH., Groeschel, S., Schultz, T. (2016). Fast and Accurate Multi-tissue Deconvolution Using SHORE and H-psd Tensors. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9902. Springer, Cham. https://doi.org/10.1007/978-3-319-46726-9_58
Download citation
DOI: https://doi.org/10.1007/978-3-319-46726-9_58
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46725-2
Online ISBN: 978-3-319-46726-9
eBook Packages: Computer ScienceComputer Science (R0)