Abstract
The frequent fluctuations in global climate variability (GCV), decreases in farmland and irrigation water, soil degradation and erosion, and increasing fertilizer costs are the significant factors in declining rice productivity, mainly in Asia and Africa. Under GCV scenarios, it is a challenging task to meet the rice food demand of the growing population. Identifying green traits (tolerance of biotic and abiotic stresses, nutrient-use efficiency, and nutritional grain quality) and stacking them in high-yielding elite genetic backgrounds is one promising approach to increase rice productivity. To this end, the Green Super Rice (GSR) breeding strategy helps to pool multi-stress-tolerance traits by stringent selection processes and to develop superior GSR cultivars within a short span of 4–5 years. In the crossing and selection process of GSR breeding, selective introgression lines (SILs) derived from sets of early backcross BC1F2 bulk populations through both target traits and non-target traits were selected. Genotyping of SILs with high-density SNP markers leads to the identification of a large number of SNP markers linked with the target green traits. The identified SILs with superior trait combinations were used for designed QTL pyramiding to combine different target green traits. The GSR breeding strategy also focused on nutrient- and water-use efficiency besides environment-friendly green features primarily to increase grain yield and income returns for resource-poor farmers. In this chapter, we have highlighted the GSR breeding strategy and QTL introgression of green traits in rice. This breeding strategy has successfully dissected many complex traits and also released several multi-stress-tolerant varieties with high grain yield and productivity in the target regions of Asia and Africa.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Green traits
- Molecular breeding strategies
- QTLs and genes
- Multiple-stress tolerance
- Green super rice varieties
1 Introduction
Food security is a global challenge for plant researchers to increase crop productivity, especially under changing climatic conditions. Rice (Oryza sativa L.) is one of the staple food crops for more than half of the world population. More than 95% of global rice is produced and consumed by the top 10 rice-producing countries (Fig. 1). China consumes about 143 million metric tons (MMT), followed by India (103 MMT), with these being the two most populated countries (https://www.statista.com). The rapid growth of population is estimated to reach 9.7 billion by 2050, and this would have a direct bearing on the demand side of global food (United Nations 2019). Worldwide, more than 800 million people are affected by malnutrition, thus hindering sustainable development programs, and food demand is expected to increase by 59–98% by 2050 (Elferink and Schierhorn 2016). Apart from this great challenge, increased global climate variability (GCV) (abiotic stresses: drought, salinity, low/high temperature, submergence/flooding; and biotic stresses: blast, bacterial leaf blight, brown planthopper, etc.) and decreasing natural resources (NRS) (e.g., decreased availability of irrigation water, labor scarcity, arable land reduction, and soil nutrient deficiency and toxicity) are the foremost factors that slow the pace toward food security for the global population. As per the prediction of the Intergovernmental Panel on Climate Change (IPCC), Earth’s global surface temperature is expected to surge by 1.4–5.8 °C by 2100 (Fahad et al. 2019), thereby resulting in a decrease in precipitation in the subtropics and a possible rise in the frequent occurrences of extreme climatic events. However, these two major factors, GCV and NRS, would have direct or indirect effects on crop growth and significantly decrease crop yield (Pandey et al. 2017). Accordingly, more than 90% of arable lands are prone to one or more than two combinations of stresses, which cause up to 70% yield losses in the major food crops (Ray et al. 2015; Fahad et al. 2017; Tigchelaar et al. 2018). To provide nutrition and food security by 2050, it has been projected that crop yield must be increased by 1%, 0.9%, and 1.6% annually for rice, wheat, and maize, respectively (Fischer and Edmeades 2010). Thus, current grain yield levels are insufficient to sustain the rapid growth of the human population. The production of the principal crops (rice, maize, and wheat) has to increase by ~42%, ~67%, and ~38%, respectively, by 2050 to meet this demand (Ray et al. 2013). As compared to cereals, rice is the only major staple food crop to have more nutritional and health benefits for increasing rice consumption; it also has sensitivity to the major biotic and abiotic stress factors. Therefore, to overcome the challenges of growing GCV and decreasing NRS, the development of superior rice cultivars with higher grain yield and multiple-stress tolerance is necessary to provide a kind of crop insurance besides increasing the income of poor farmers across mainly Asian and African countries.
Multiple abiotic stresses greatly influence crop productivity and have adverse effects on plant growth and development (Fahad et al. 2017; Cohen and Leach 2019). For example, drought is a major constraint in rice production and approximately 42 million hectares of rice in Asia suffer significant yield losses from varying amounts of drought stress at the different crop growth stages (Saikumar et al. 2016; Sandhu and Kumar 2017; Mukamuhirwa et al. 2019). Besides drought, soil salinity and alkalinity are also devastating factors that cause significant yield losses of rice crops at the vegetative and reproductive stages. Globally, about 20% of the cultivable land and 33% of irrigated agricultural areas are afflicted by salinity stress (Shrivastava and Kumar 2015). Thus, it is crucial to identify stress-tolerant rice varieties to maintain productivity and meet global food security needs. Beyond abiotic stresses, sustainable rice production has become one of the top priorities in developing and adopting eco-friendly rice varieties with low input-use efficiency. In the majority of the developed countries, farmers have been using considerably more fertilizer to increase their crop yield. According to Cheng et al. (2007), rice production in China has to be increased by 14% by 2030 to meet the food requirements of its rapidly growing population. In Jiangsu Province, farmers are applying 300–350 kg N/ha to achieve high yield for rice crops (Sui et al. 2013), which is 90% more than the global average N application (Chen et al. 2014a) . The excess amounts of nutrient fertilizer also cause the accumulation of higher salt concentration in the soil. This further decreases water and nutrient absorption, leading to several changes in physiological processes as leaf dehydration decreases photosynthesis efficiency, decreases grain micronutrients, and also causes severe environmental pollution (Kreuzwieser and Gessler 2010; Panda et al. 2012; Guo et al. 2017; Ali et al. 2018b). Even in China, the excessive amounts of fertilizer application in paddy lands to boost yield are no longer a viable option, especially under increasing fertilizer costs. This is further driving the search for rice cultivars with high nutrient-use efficiency that are required for sustainable rice production and increasing grain yield under optimal and suboptimal rates of fertilizer dosage. Thus, it is essential to develop rice varieties with high nutrient-use efficiency to maximize yield and productivity with the best agronomic management options such as judicious stage-specific dosages of fertilizer application. Further, it is essential to develop high-yielding, multiple-stress-tolerant rice varieties with combinations of several green traits such as tolerance of/resistance to drought, salinity, high/low temperature, flooding, blast, bacterial blight, tungro, brown planthopper, and stem borer, along with water-use efficiency. These multi-stress-tolerant varieties also need to meet market segment requirements such as duration, grain shape, and quality preferences. Recently, the incorporation of desirable nutrients for improved grain quality such as iron and zinc in multi-stress-tolerant cultivars has become necessary in rice improvement programs to ensure food security and overcome hidden hunger (Ali et al. 2020; Yu et al. 2020).
2 Green Super Rice
During the past two decades, breeders and biotechnologists have been working on various biotic and abiotic stress tolerances with a focus on increasing crop yield. This is possible by modifying the plant architecture and introgressing the target traits into the desired background through conventional and marker-assisted breeding approaches (Mehta et al. 2019; Oladosu et al. 2019; Gautam et al. 2020; Muthu et al. 2020). Complex abiotic stress tolerances such as drought and salinity tolerance are polygenic, and may have a negative association with grain yield components and might also interact with genetic and physiological mechanisms of similar traits (Guojun et al. 2009). Also, choosing the right donors for different target traits is challenging. In most cases, the abiotic stress-tolerant rice varieties are developed by crossing with tolerant landraces. However, concern exists regarding the difficulty in breaking undesirable linkages through conventional breeding approaches, which often represent a time-consuming process for selecting and fixing desirable lines, with limited success (Zhou et al. 2010; Wang et al. 2013). These developed tolerant rice varieties are low to moderate yielding under irrigated and rainfed conditions. In light of this apprehension, the Green Super Rice (GSR) breeding strategy began in 2008 at IRRI to efficiently develop lines with multiple-stress tolerance and more nutrient- and water-use efficiency with high genetic gains for various targeted ecosystems. The development of superior rice varieties with the GSR breeding approach under decreased rates of fertilizer, pesticide, and irrigation water, grown in marginal environments and producing fewer greenhouse gas emissions, and with improved grain nutrient elements (Zhang 2007; Wing et al. 2018), is described in Fig. 2. GSR breeding aims to develop stable high-yielding GSR rice varieties with several green traits suitable to be grown under lower input conditions in irrigated and rainfed areas of Asian and African countries.
A schematic representation of the Green Super Rice (GSR) breeding program for the development of green traits and increasing grain yield with improved grain quality. (a) The drastic changes in climatic variations, degradation of natural resources, and increasing global population are the major driving forces for increasing food demand by about 42% by 2050. (b) To ensure food security against these challenges, the GSR concept was proposed in 2005, and its major focus was to develop novel rice varieties with green traits, which included tolerance of multiple stresses (biotic and abiotic), high nutrient-use efficiency, fewer inputs of fertilizer, and water-saving. The combination of green traits and the GSR breeding concept provided an environment-friendly and natural resource–saving dimension. (c) The major target of GSR breeding is to develop new GSR cultivars through the integration of an advanced genomics platform for determining the genomic regions for green traits and further disseminating promising cultivars for specific target regions to improve the income of resource-poor farmers. (d) As of now, many QTLs have been identified using the GSR breeding strategy, and several genes have been functionally characterized and cloned for increasing the tolerance of abiotic stresses (Ali et al. 2020; Yu et al. 2020). These identified QTLs and genes provide abundant genetic information for the development of novel GSR varieties
Integrating advanced genomics and stringent phenotypic selection in the GSR breeding strategy helped to identify promising rice varieties besides understanding the molecular genetics and physiological mechanisms underlying trait expression (Wing et al. 2018; Ali et al. 2020; Yu et al. 2020). This breeding strategy played a significant role in the quick introgression of the desired target traits with the highest precision and with less or no genetic drag.
2.1 GSR Breeding and Population Development
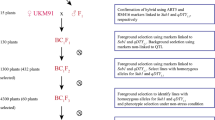
The concept of GSR and its breeding strategy followed a systematic breeding effort and advanced genotyping approaches for developing varieties with significantly improved tolerance of multiple biotic and abiotic stresses, along with improved nutrient- and water-use efficiency. A schematic view of the backcross GSR breeding technology for developing early backcross-selective introgression lines (EB-SILs) appears in Fig. 3. The development of several promising stress-tolerant rice cultivars that withstand drought, salinity, and flooding through a selective introgression breeding approach started at IRRI in 1998 (Li et al. 2005; Ali et al. 2006; Lafitte et al. 2006). These early efforts helped in the development of the GSR breeding strategy to breed varieties with multiple biotic and abiotic stress tolerance. Starting in 2009, IRRI focused on developing cultivars with multiple abiotic stress tolerance through an innovative GSR breeding strategy to benefit poor and smallholder farmers (Ali et al. 2012). The concept of a GSR breeding program primarily involved two fundamental steps: first, developing superior EB-SILs; second, identifying suitable SILs to develop designed QTL pyramiding (DQP) lines. These two approaches helped to develop GSR varieties with multiple biotic and abiotic stress tolerance without compromising on grain yield, grain appearance, and cooking quality parameters.
IRRI-Green Super Rice early backcross breeding strategy to develop rice varieties with multiple-stress tolerance, resource-use efficiency, and high yield with market-required grain quality. (a) The selected donors were crossed with HHZ and WTR-1 and later selfed to create 16 BC1F2 bulk populations. These backcrossed lines were used in three rounds of phenotypic selection under different environments and selected lines with better performance were evaluated in the different targeted ecosystems. (b) Identified promising trait-specific selective introgression lines with more than one target green trait were used for the designed QTL pyramiding to combine various desirable traits into one single line. The blue-colored circles indicated in the schematic diagram were used to show the different genotyping technologies used to identify genomic regions associated with green traits in GSR breeding populations. (IRG, irrigated conditions; DSF, drought stress in field conditions; BDS, biotic-stress phenotypic screening; RFD, rainfed environment; SAL, salinity at the seedling stage at 18 d/sm (deciSiemens per metre); COL, cold stress; LIP, low inputs (without any fertilizer, pesticide, or herbicide); ARG, anaerobic germination (direct-seeded and immediately submerged in water for 21 days and maintained at 10-cm water depth); SBM, submergence for 21 days at 14 days of seedling stage
The GSR breeding program at IRRI initially focused on identifying and exploring genetic variation and further selecting ~500 elite EB-SILs in an elite and widely adaptable Chinese variety, Huang-Hua-Zhan (HHZ), background with 16 donors from the mini-core collection of rice germplasm and a part of the 3000 Rice Genomes Project. These parental varieties underwent whole-genome sequencing to identify genomic variations. In the IRRI GSR breeding program, two additional elite recipients, Weed Tolerant Rice-1 (WTR-1) and TME80518, were used for developing EB-SILs (Ali et al. 2012, 2013b, 2018a). In this GSR breeding program, the BC1F2 populations were derived from crosses between the recipients (HHZ, WTR-1, and TME80518) and the 16 donors at IRRI. The F1s derived from the HHZ background were again backcrossed with HHZ and subsequently selfed, and the progenies bulked to generate a total of 16 BC1F2 bulk populations. These bulk populations were screened over three rounds of selection for different abiotic stresses (drought, salinity, submergence, and low-input fertilizer) and biotic stresses (blast, bacterial leaf blight, and tungro), and in normal irrigated conditions. This led to the identification of a total of 1333 trait-specific SILs from the HHZ background, 2232 SILs from the WTR-1 background, and 1408 SILs from the TME80518 background.
The developed SILs were phenotyped across different stress and non-stress conditions and genotyped with SSR and high-density SNP markers. The phenotypic and marker data generated using SSR and SNP analysis allowed us to identify the donor QTLs associated with target abiotic and biotic stress tolerance/resistance and yield in the SILs. Then, the best SILs were selected as parents based on both their superior phenotypes and complementary donor alleles for designed QTL pyramiding (DQP) to develop better GSR varieties that combine various target traits from two or more donors. These promising SILs showed high tolerance vis-à-vis the respective tolerant checks IR74371-70-1-1, FL478, IR49830, and PSBRc82 under normal and stress conditions. Through these DQP approaches, a total of 2023 pyramiding lines (PDLs) from the HHZ background and 661 PDLs from the WTR-1 background were developed and found superior to the checks for all of the traits studied (Ali et al. 2013a, 2020; Li and Ali 2017). The selected 564 EB-SILs derived from the genetic background of WTR-1 as recipient parents and 11 donor parents were sequenced using tunable genotyping-by-sequencing (tGBS) technology. Table 1 provides the list of donors and recipients used. These SILs developed through early backcross breeding were more advantageous in terms of high allelic diversity, which comprised multiple donor parents. As compared with the biparental mapping population, these SILs provide more precise QTL detection and fine-mapping of candidate genes for biotic and abiotic stress tolerance (Ali et al. 2017, 2018a). A total of 102 loci were identified from the 11 populations, and they contain 120 deleterious SNPs, with a range of one to four SNPs per loci. These loci can substitute the amino acid, and this leads to changes in the functional proteins in either positive or negative regulations (Ali et al. 2018a). Interestingly, the donors of SILs have a different introgression frequency of alleles in the recipient genomic regions and are mainly attributed to the severe selection pressure under abiotic stress conditions. With the availability of tGBS marker information with the GSR breeding approach, several genetic loci have been identified through the various linkage-based mapping methods. This innovative approach assured the development of new rice varieties that were tolerant of multiple stresses.
3 Genetics of Green Traits
In the GSR breeding program, the critical genetic determinants of various abiotic stress tolerances can be identified through QTL mapping approaches. As of now, the details of the identified QTLs and breeding materials are listed in Table 2. In many instances, advancement in the identification of stress-tolerance genes and QTLs that determine the trait has led to insights into the specific physiological and molecular mechanisms in response to various biotic and abiotic stresses (Wing et al. 2018; Yu et al. 2020). The highlights of the green traits associated with genetic information that confers stress tolerance and adaptive strategies provide valuable information for crop improvement.
3.1 Drought Tolerance
Drought is a complex trait. Understanding its mechanisms in terms of plant-water relationship traits, molecular breeding strategies, and dissecting the molecular genetics of QTLs and deployment in the breeding pipeline are the key steps for the development of drought-tolerant rice varieties. For drought improvement, it is essential to use advanced genomics and omics platforms that provide an opportunity for the mining of trait-specific allele functions. Identifying and developing drought-tolerant rice cultivars mainly depends on two major factors: prospecting for donors and effective phenotypic evaluation methods required for crossing programs and a successful selection scheme. However, improving drought tolerance in rice varieties has had slow progress due to the complexity of the trait and some associated undesirable characteristics, including low grain yield and a lack of preferable nutritional grain quality traits (Swamy and Kumar 2013; Kumar et al. 2014). With pre-breeding activity and marker-assisted breeding approaches, several drought-tolerant lines have been developed and further used in breeding programs for improving lines with multiple-stress tolerance (Singh et al. 2016; Ali et al. 2017; Pang et al. 2017a; Gautam et al. 2020; Kumar et al. 2020). However, improving yield potential under drought environments is limited due to the intensity, duration, and timing of drought stress. Important are the establishment of efficient phenotypic screening protocols and using precise phenotypic selection criteria (Ouk et al. 2006; Oladosu et al. 2019). From the recent developments in various molecular breeding strategies, precise high-throughput phenotypic technologies have been identified for promising traits such as grain yield, and morpho-physiological component traits have shown moderate to high heritability under drought stress conditions (Tuberosa 2012; Kumar et al. 2014; Swain et al. 2014; Ali et al. 2017; Sahebi et al. 2018; Yadav et al. 2019). Several researchers have used direct selection of GY, along with considering other secondary traits using molecular and genomic approaches (Oladosu et al. 2019). The results led to the identification of the QTLs and genes that are associated with these traits. Discovering these QTLs/genes that are responsible for tolerance traits is essential for developing crops with tolerance through marker-assisted selection or genetic engineering approaches. To this end were identified the major-effect drought tolerance QTLs from drought-tolerant rice varieties Apo (qDTY1.1, qDTY2.1, qDTY3.1, and qDTY6.1), Way Rarem (qDTY12.1), Nagina 22 and Dhagaddeshi (qDTY1.1), and Vandana (qDTY2.3 and qDTY3.2) (Bernier et al. 2007; Venuprasad et al. 2009; Vikram et al. 2011, 2015; Ghimire et al. 2012; Dixit et al. 2014). With the marker-assisted introgression approach, grain yield QTLs with major and consistent effects (qDTY1.1, qDTY2.1, qDTY2.2, qDTY3.1, qDTY3.2, qDTY6.1, and qDTY12.1) on chromosomes 1, 2, 3, 6, and 12 have been validated, deployed into drought breeding programs, and successfully improved drought-susceptible rice mega-varieties (Kumar et al. 2014, 2017b, 2020; Singh et al. 2016; Sandhu and Kumar 2017). However, the locally adapted high-yielding commercial mega-varieties show a sensitive reaction to other biotic and abiotic stresses. These improved tolerant rice varieties have not been able to show a similar yield performance in varied agroecosystems. Introgression of these major QTLs/genes in a different combination using a marker-assisted selection approach provides an opportunity to increase desirable phenotypic traits and thereby improve grain yield. Still, pyramiding of these major QTLs/genes has not been fully exploited due to G × E interaction of this complex trait epistasis, and pleiotropy could have negative/positive effects on the trait of interest (Li 1998; Yano et al. 2003; Xu and Crouch 2008). As a consequence, it has been tough to make significant genetic improvements in grain yield under drought stress through conventional breeding methods. Therefore, the development of large-scale phenotypic screening, marker-assisted breeding strategies, and advanced genotyping technologies has significantly accelerated varietal improvement programs on drought tolerance. Further, the identification of QTLs/genes affecting grain yield could result in yield improvement and stability under drought stress. In comparison to the classical breeding and genetic mapping approaches, the GSR breeding strategy used SILs and designed QTL pyramiding approaches in the development of multiple-stress-tolerant rice varieties for improving grain yield under stress conditions.
The GSR breeding strategy has three significant advantages: first, the selected few SILs require lesser costs in both genotyping and phenotyping; second, the strategy significantly increased the power in detecting the genomic regions of QTLs/genes; third, the selected lines mostly carried the beneficial alleles of QTLs (Ali et al. 2017). Based on the DQP approach, Feng et al. (2018) identified a total of nine drought tolerance QTLs on chromosomes 2, 3, 5, 6, 8, and 12 using a segregation distortion approach. These QTLs were detected from the three different genetic backgrounds of trait-specific SILs, which were generated from the 63 SILs from the cross HHZ × Teqing, 68 SILs from the cross HHZ × CDR22, and 75 SILs from the cross HHZ × OM1723 (Ali et al. 2017). The major significant QTLs on chromosomes 3, 5, and 8 were closely associated with the earlier reports of grain yield QTLs and seed fertility QTLs under drought stress. Similarly, researchers exploited the genotypic information on EB-SILs derived from the 11 donors crossed with recurrent parent WTR-1, which comprised a total of 564 diverse SILs used for identifying synonymous and non-synonymous deleterious polymorphic SNPs (Ali et al. 2018b). Of these, the significant locus Os01g01689 on chromosome 1 possessed a G/A deleterious SNP altering an amino acid from Ala to Thr in the background of Haoannong and Y-134. This locus was associated with abiotic stress tolerance. This innovative breeding strategy addresses the GCV challenge and allows poor farmers to benefit from the use of stress-tolerant and high-yielding rice varieties under limited resources. Therefore, genotyping the EB-SILs and DQP populations helped to identify several promising genomic regions for complex traits and assisted in molecular marker development and genomic applications in molecular breeding programs.
3.2 Salinity Tolerance
More than 20% of global agricultural land is affected by salinity, especially in the coastal regions of South and Southeast Asian countries. Salinity is mainly caused by the drastic changes in the climatic events that are significantly associated with increases in salinity in the soil. During crop growth stages, the accumulated Na+ and Cl− concentrations in plants lead to inhibiting cell expansion and photosynthetic activity, followed by changes in physiological and molecular pathways in roots for the uptake of water and nutrients, leading to cytotoxic effects. Mainly, the accumulation of these elements can be processed through osmotic and ionic stresses, which can activate the formation of reactive oxygen species (ROS) and cause leaf damage or plant death. Thus, developing rice varieties with salinity tolerance is a prominent approach to resolve the salinity problem. However, progress in developing salinity-tolerant rice varieties has been slow because of the complexity of the salinity tolerance trait, and this has involved multiple physiological and biochemical pathways. As of now, several researchers had worked on the complex traits using advanced genomic-assisted breeding tools in traditional molecular breeding strategies and sequencing at the whole-genome level. In the past two decades, >700 QTLs and 200 genes have been reported for salinity tolerance at different growth stages (Rahman et al. 2017; van Oort 2018) (http://archive.gramene.org/). Most of these genes and QTLs are associated with different traits such as salt injury score, shoot and root growth, fresh and dry weight of shoot and root, Na+ and K+ content in shoot and root, grain yield, and chlorophyll content. The genomic regions of these QTLs and genes are involved in the different physiological and molecular mechanisms such as ionic equilibrium, osmotic adjustment, transcription regulation, and signaling pathway (Molla et al. 2015; Pradhan et al. 2015; Gimhani et al. 2016; Kumar et al. 2017a). However, the identified genomic regions have a more significant interval gap because they employ a low density of molecular markers and possibly because of epistatic QTLs and environmental interaction. It’s possible to have hundreds of genes in those QTL positions, which makes it difficult to track the genetic pathways.
The recent development of genotyping technologies provided robust genotyping information and can remarkably reduce chromosomal intervals and also help in identifying accurately predicted gene functions related to the target traits. Dissecting these complex traits and identifying superior tolerant lines, the GSR breeding program began an integrated molecular breeding strategy to identify genome-wide trait-specific introgression lines (ILs) through the DQP approach. Using high-density SNP markers of tGBS technology, three significant QTLs, qSES2, qSES4 (qChlo4), and qChlo1, were mapped, and this further narrowed down the list of genes through the use of grandparent genotypic information (Pang et al. 2017b). As a result of this approach, 13, 34, and 40 candidate genes were identified in the QTLs regions, respectively. Pang et al. (2017b) carried out functional analyses of the candidate genes for the salinity-tolerance QTLs to infer two, five, and six genes as the most likely candidates of qSES2, qSES4 (qChlo4), and qChlo1, respectively. This combination of high-density SNP markers with parent and grandparent information could be a potential approach for identifying the right genes for desired target traits in rice. With this strategy, several high-yielding, multi-stress-tolerant rice varieties such as NSIC Rc480, NSIC Rc534, NSIC Rc 390, NSIC Rc 392, NSIC Rc 554, and NSIC Rc 556 were developed and released for cultivation under saline conditions (Guan et al. 2010; Ali et al. 2017; Yu et al. 2020).
3.3 Submergence Tolerance
Submergence is an essential factor that limits rice yield over 15 million ha in rainfed lowland regions of Asia (Septiningsih et al. 2009). Rice is known to adapt well to flooded conditions, but most rice cultivars become vulnerable upon complete submergence. Complete submergence of rice plants, particularly under turbid water, causes severe damage through decreased respiration and photosynthesis (Ella et al. 2003; Bailey-Serres et al. 2010). Submergence tolerance is an important trait that could help rice plants to overcome flooding with a water head of 1 m for 14 days at the active tillering stage. The genetics of submergence tolerance from FR13A showed a high heritability and was governed by both major genes and some quantitative trait loci (QTLs) (Suprihatno and Coffman 1981; Nandi et al. 1997; Sripongpangkul et al. 2000; Toojinda et al. 2003). Xu et al. (2006) used FR13A for cloning Sub1A, a major QTL on chromosome 9, which is a transcription factor concerning ethylene responsiveness. Sub1A-1, one of the alleles at Sub1A, was found to be essential for submrgence tolerance in several semi-dwarf varieties (Septiningsih et al. 2009). Ali et al. (2006) demonstrated that progenies with submergence tolerance were identified consistently in most BC populations derived from sensitive parents. This suggested the presence of hidden genetic diversity for submergence tolerance in the primary gene pool of rice. Wang et al. (2015) determined the genetic basis of submergence tolerance in rice and facilitated the simultaneous improvement of submergence tolerance in rice. In their study, they characterized the genome-wide responses of 162 SILs with submergence tolerance from 12 BC populations using SSR markers that helped in the dissection of the hidden diversity and transgressive segregation. Their results provided insights into the genetic basis of submergence tolerance of rice and demonstrated a novel strategy for simultaneous improvement and genetic dissection of complex traits using the approach of selective introgression (Li et al. 2005). The genome-wide responses of donor alleles to strong phenotypic selection for submergence tolerance can be understood with three key findings from Wang et al. (2015). First, they found significant over-introgression of the donor alleles at 295 loci in 167 functional genetic units (FGUs) across the rice genome. Second, they observed significantly increased homozygosity or “loss of heterozygosity” genome-wide. Third, pronounced non-random associations between or among the detected submergence tolerance loci led to the discovery of putative genetic networks (multi-loci structures) underlying submergence tolerance in rice. Further, their results suggested that submergence tolerance of rice is controlled by large numbers of loci involved in multiple positively regulated signaling pathways (Wang et al. 2015). It is essential here to understand that the restoration of one or more of these broken pathways in the BC progenies by genetic complementation from the introgressed functional donor alleles at submergence tolerance loci provided an appropriate explanation for the transgressive segregation of submergence tolerance and this could be extended to other complex traits in rice. The GSR breeding strategy developed several promising salinity-tolerant materials such as NSIC Rc480, GSR 5, and GSR11. Yorobe et al. (2016) found that the mean difference in net farm income between GSR and non-GSR varieties was positive and significant and gave farmers an income advantage of USD 230.90/ha for GSR variety users. In their study, Yorobe et al. (2016) found that, with a high occurrence of flooding in the wet season, the use of GSR varieties assured rice farmers of a positive net farm income.
3.4 Nutrient-Use Efficiency
Nutrient-use efficiency (NUE) is one of the most critical traits for increasing yield and productivity by using nutrients such as nitrogen (N), phosphorus (P), and potassium (K) to increase NUE. During the Green Revolution (GR) period in the 1960s and post-GR, yield increased significantly, and this was primarily achieved with a higher input dosage of fertilizer, pesticide, and water (Ali et al. 2018b). Recently, Hawkesford and Griffiths (2019) mentioned that only 33% of global N is recovered from harvested grain, while the remaining N is a significant pollutant and a colossal waste of resources. A higher amount of fertilizer application can lead to a substantial imbalance in nutrient availability in the soil, increase the risk of pests and diseases, and not be cost-effective for poor farmers (Chen et al. 2014a, b; Rahman and Zhang 2018). Regarding environmental safety and the use of low inputs in farmers’ fields, the GSR breeding team started a program at IRRI to identify nutrient-use-efficient breeding lines that could obtain higher grain yield under integrated nutrient management techniques. This was the first initiative in the NUE breeding program at IRRI (Jewel et al. 2018) and it was reported as a unique and systematic breeding approach through the selection of SILs with higher NUE through the early backcross breeding program. SILs were selected in four consecutive seasons under different nutrient fertilizer combinations of N, P, and K dosages. Five promising SILs (Nue-115, Nue-114, Nue-112, Nue-229, and Nue-230) were identified with higher grain yield and nutrient-use efficiency. These SILs could provide valuable information for rice breeding programs. The genetics of NUE (Mahender et al. 2019) identified a total of 19 QTLs that were associated with three agronomic traits by using tGBS technology. These major QTLs were located on chromosomes 2, 5, 8, 9, and 12. The genomic regions of these QTLs were co-localized with earlier reports of low nitrogen and phosphorus conditions. Importantly, in silico analysis of these QTL positions suggested that several key candidate genes played a major role in the various molecular and physiological pathways in response to abiotic stress tolerance and also maintenance of the homeostasis mechanism under low-input conditions. Similarly, Jewel et al. (2019) detected a total of 49 main-effect QTLs under six nutrient conditions. These QTLs explained a phenotypic variance range from 20.25 to 34.68%. They were located on all 12 chromosomes, except on chromosomes 7, 11, and 12. Among them, four hotspot QTLs were identified on chromosomes 3, 5, 9, and 11. Interestingly, 22 QTLs for partial factor productivity and four QTLs for agronomic efficiency were detected as novel under –P and 75 N conditions. Several genes and transporters were located in the interval regions of these QTLs, and they were involved in nutrient uptake and transporting mechanisms from soil to plants. Therefore, the hotspot regions of QTLs and genes may offer significant value for marker-assisted selection and pyramiding of multiple QTLs for improving NUE in rice.
3.5 Weed-Competitive Ability Traits
Threats such as GCV, rising labor shortages, decreasing arable land, and the increasing prices of fertilizer and pesticide are the major contributors to the decrease in rice production (Singh et al. 2013). To overcome these constraints, shifting from the manual rice transplanting system to direct-seeded rice (DSR) is the most promising approach to improve rice sustainability (Chauhan and Abugho 2013; Mahender et al. 2015). It has numerous benefits, mainly in decreasing water use by 35–75%, decreasing labor demand, shortening crop duration, mitigating methane gas emissions, and lowering the cost of cultivation (Mahender et al. 2015; Dimaano et al. 2017). However, vigorous growth of weeds is one of the most complicated biological constraints in the DSR system to attaining optimal grain yield (Antralina et al. 2015; Chauhan et al. 2015a, b; Jabran and Chauhan 2015). Several options such as tillage operations and herbicide application are available to control weeds, but these are laborious and costly (Rahman et al. 2012). Globally, more than USD 100 billion are lost annually because of weed control (Appleby et al. 2002). Therefore, developing a breeding strategy for weed-competitive (WC) rice varieties is a critical solution for decreasing tillage operations, hand weeding, and herbicide inputs in the DSR system. WC ability is a complex trait, and it interacts with several agro-morphological traits (Chauhan et al. 2015a; Raj and Syriac 2017). Thus, it is essential to understand the trait interactions and mechanisms that confer WC ability, and this could be useful for speeding up the breeding activities for developing WC rice cultivars. Recently, as a part of the GSR breeding program at IRRI, Chauhan et al. (2015b) and Dimaano et al. (2017) followed a systematic breeding effort to identify WC ability traits related to early seed germination (ESG) and early seedling vigor (ESV). The breeding materials were developed from the four early generations of backcross populations derived from one common recipient parent, Weed Tolerant Rice-1 (WTR-1), and four donors, Y134, Zhong 143, Khazar, and Cheng Hui-448. These SILs were evaluated over three rounds of selection in upland weed-free, upland weedy, and lowland weedy conditions. Among the total SILs, five (G-6-L2-WL-3, G-6-RF6-WL-3, G-6-L15-WU-1, G-6-Y16-WL-2, and G-6-L6-WU-3) were found to be promising in lowland weedy conditions, whereas four SILs (G-6-Y7-WL-3, G-6-Y6-WU-3, G-6-Y3-WL-3, and G-6-Y8-WU-1) were found to have the highest grain yield under upland weedy conditions (Dimaano et al. 2017). The primary requirements for the DSR system to be successful are the following: uniformity and speed of germination rate and early seedling growth are significantly associated with robust and vigorous crop growth and a better crop establishment, which can influence WC ability (Cui et al. 2002; Foolad et al. 2007; Diwan et al. 2013; Dang et al. 2014). These traits can provide support for traits attributed to efficient root growth that can help in the absorption of more nutrients (Farooq et al. 2011; Matsushima and Sakagami 2013; Singh et al. 2015; Khan et al. 2016). For the molecular genetics of WC rice cultivars, a total of 43 QTLs were mapped on all 12 chromosomes, except on chromosomes 4 and 8, by using 677 high-quality SNP markers (Dimaano et al. 2020). Interestingly, 29 novel genetic loci were associated with ESG and ESV traits on chromosomes 1, 3, 5, 6, 7, 10, 11, and 12. The hotspot regions of chromosomes 11 and 12 were associated with multiple traits (Dimaano et al. 2020). Many of these QTLs were co-localized in previous reports, which are related to germination rate, germination index, germination percentage, and germination time in different genetic backgrounds of mapping populations (Mahender et al. 2015). In addition, some critical genes located in the co-localized hotspot regions can influence the regulation of various physiological functions such as chloroplast development, photosynthesis, hybrid sterility, seed development, and seedling lethality during plant growth stages (Gothandam et al. 2005; Matthus et al. 2015; Sharma and Pandey 2016; Yu et al. 2016). Therefore, the hotspot regions with co-localized QTLs and genes may have a more significant role in the improvement of weed-competitiveness, mainly in African and Asian countries, to decrease rice production costs.
3.6 Low-Temperature Stress Tolerance at Different Crop Growth Stages
Rice is one of the most sensitive among the cereal crops to low-temperature stress (LTS)/cold stress (CS), mainly at the germination to booting stage, which can cause a significant yield decrease due to reduced germination rate and seedling growth, high spikelet sterility, delay in flowering, and lower grain filling (Ranawake et al. 2014; Schläppi et al. 2017; Shakiba et al. 2017; Najeeb et al. 2020). The drastic changes in GCV in rice-growing areas mainly in the tropical, subtropical, and temperate regions had severe effects from low-temperature stress during the critical stages of seedling growth and pollen abortion at the booting stage, leading to grain yield decreases (Ye et al. 2009; Jena et al. 2010; Sun et al. 2018). LTS mainly inhibits sugar accumulation in the pollen and further leads to male sterility, which is regulated by the invertase enzyme through the hormone abscisic acid (ABA) pathways that transport sugar to the tapetum. The results of these mechanisms decrease invertase amounts in susceptible rice varieties, leading to lower pollination (Oliver et al. 2007). This situation warrants the development of LTS-tolerant varieties through a systematic breeding effort to increase rice production in 25 countries, including the major rice-producing countries (Cruz et al. 2013; Zhang et al. 2017). Therefore, to minimize yield losses under LTS, particularly in cold-affected regions, it is crucial to identify potential donors to improve LTS tolerance. LTS tolerance breeding for widely adapted and high-yielding rice cultivars is needed to meet future food demand worldwide.
Information on the genomic regions of QTLs and genes governing tolerance of LTS is limited for different growth stage-specific traits in rice due to its complex nature, and this significantly influences QTL × environment interactions. Up to now, several studies have been reported for QTL mapping for LTS. Recently, Najeeb et al. (2020) reviewed the genetics of LTS tolerance QTLs in rice. A total of 239 and 339 QTLs were identified on 12 chromosomes using genome-wide association studies and biparental mapping populations, respectively (http://archive.gramene.org). However, mapping of the sensitive stage, especially at the reproductive phase of a complex trait, involves multiple genes, environment interactions, and difficulties in phenotypic screening. Despite these large numbers of QTLs for LTS, a few of them were studied for fine-mapping and cloning. Based on the public domain and rice database, a total of 38 candidate genes were functionally characterized for LTS tolerance. Importantly, four genes on chromosome 1 (OsCOIN, OsGSK1, OsGH3-2, and OsMYB3R-2), two genes on chromosome 6 (OsiSAP8 and OsbZIP52,) and a single gene on chromosome 4 (OsCAF1B) and chromosome 11 (OsAsr1) were significantly associated with LTS tolerance in the seedling and reproductive stage in rice. In contrast, breeding strategies for LTS tolerance remained a slow process by conventional methods because most of the QTL mapping methods could not explain G × E interaction well and appropriate statistical and advanced breeding strategies were lacking. Improvement of LTS tolerance at the reproductive stage is needed through a selective introgression method (Liang et al. 2018). A population was derived from BC2F4 onward using five donors and a japonica (Geng) recipient parent that were screened over three rounds to select under LTS conditions at the reproductive stage. This approach helped in dissecting the complex trait of LTS tolerance and developed trait-specific introgression lines. A total of 17 QTLs were identified using five different populations of EB-SIL derived from five different donors into a common recipient parent. Further, multi-locus probability tests and linkage disequilibrium results showed that a total of 46 functional genetic units were distributed across the rice genome for cold tolerance. Studies showed the presence of strong epistasis and power of the statistical approach for the development of selective introgressions for simultaneous improvement and genetic dissection of complex traits. Zhu et al. (2015) used an inter-connected breeding (IB) population comprising 497 SILs derived from eight BC families with the same recipient parent for identifying and fine-mapping QTLs for LTS tolerance at the booting stage. A total of 41,754 high-quality SNPs were obtained through the re-sequencing of the IB population. Phenotyping was conducted under field conditions in 2 years and three locations. Association analysis identified six QTLs for LTS tolerance on chromosomes 3, 4, and 12. The stably expressed QTL qCT-3-2 was fine-mapped and narrowed down to approximately 192.9 kb on the reference genome (Zhu et al. 2015). The QTL qCT-3-2 is essential for developing varieties with LTS tolerance at the booting stage, which are in high demand in temperate and high-altitude rice production regions. GWAS applied to an IB population allowed better integration of gene discovery and breeding. QTLs can be mapped in high resolution and quickly used for breeding.
3.7 Grain Quality
Improvement of grain quality traits (grain appearance, cooking and eating quality) is a major concern in rice breeding programs. Earlier rice breeding efforts over the past few decades primarily focused on increasing grain yield potential as the primary target in the major Asian rice-growing countries, and most of the high-yielding and popular rice varieties had poor grain quality traits (Custodio et al. 2019). Approximately one-third of the global population is suffering from nutritional deficiency, mainly caused by inferior grain quality traits such as low protein content and lack of vitamins and minerals (Balyan et al. 2013). Therefore, to meet the global food demand, it is essential to keep nutritional quality traits integrated well in our breeding programs. Consumer acceptance and their preferred grain traits are major factors that influence different markets across the globe. For instance, grain appearance traits such as long and slender grain shape are mostly preferred in countries such as India, Vietnam, the United States, and most Asian countries, whereas northern China, Korea, and Japan prefer short and round rice grains (Unnevehr et al. 1985; Cuevas et al. 2016). Improvement in grain quality is for a complex trait involving many traits such as milling efficiency, grain appearance (size and shape), and cooking and eating quality (apparent amylose content, gelatinization temperature, gel consistency). Many of these grain quality component traits are highly complex and quantitative in nature and are significantly influenced by environmental factors. In a global view, grain shape traits need to be understood, matching market and consumer preferences and needing to be adequately addressed in breeding programs for different target regions. Several researchers have identified many critical genetic loci and candidate genes for grain quality traits (Yun et al. 2014; Balakrishnan et al. 2016; Pang et al. 2016; Wang et al. 2017; Mogga et al. 2018; Kavurikalpana and Shashidhara 2018; Wing et al. 2018; Bazrkar-khatibani et al. 2019; Calayugan et al. 2020; Zhang et al. 2020). Mahender et al. (2016) reviewed grain nutritional traits and associated genetic information on QTLs and genes in rice. More than 400 QTLs have been reported for these grain quality traits, including grain appearance and cooking and nutritional properties (Mahender et al. 2016; Bazrkar-khatibani et al. 2019).
As of now, 28 major genes are involved in controlling grain shape and 65 genes are associated with eating quality traits in rice (http://qtaro.abr.affrc.go.jp/). Importantly, the natural variation of eating quality genes such as ALK and WAXY on chromosome 6, Badh-2 on chromosome 8, and LOX-3 on chromosome 3 regulates cooking quality traits and encodes the strong component in rice fragrance (Chen et al. 2008; Shirasawa et al. 2008; Gao et al. 2011; Venu et al. 2011; Wang et al. 2017). Similarly, overexpression of the six genes (OASA2, BiP, OASA1D, OsAAT1, OsAAT2, OsISA1, and OsISA2) on chromosomes 1, 2, 3, 5, and 8 significantly increased amino acid content and seed storage protein content in rice grain (Zhou et al. 2009; Saika et al. 2011; Utsumi et al. 2011). Recent successful GSR breeding strategies help to a great extent to understand the various green traits that include grain quality and biotic and abiotic stress tolerance genes that have been cloned (Yu et al. 2020). These include the critical genes OsAAP6 on chromosome 1, OsGRF4 on chromosome 2, three genes (GNP1, lgy3, and GL3) on chromosome 3, two genes (Chalk5 and GW5) on chromosome 5, two genes (W7 and OsSPL13) on chromosome 7, and a single gene (OsOTUB1) on chromosome 8 associated with grain quality traits, and some of these genes are significantly associated with yield-attributed traits and NUE traits (Li et al. 2018). Recently, Mahender et al. (2019) identified 19 QTLs for agro-morphological traits under different dosages of nutrient fertilizer. Interestingly, one of the major QTLs for leaf chlorophyll content on chromosome 2 and in the same genomic region co-localized with the nitrate transporter OsNPF7.2 and GROWTH-REGULATOR FACTOR 4 (OsGRF4). These hotspot genomic regions play a vital role in the interaction of GRF4 and DELLA proteins that are involved in multiple gene regulation related to grain quality traits and that also maintain homeostatic coordination of nitrogen and carbon metabolism (Serrano-Mislata et al. 2017; Li et al. 2018; Xing et al. 2018).
3.8 Biotic Stress Tolerance
Rice grain yield declined globally by more than 52% because of various biotic stresses such as insect pests (brown planthopper, green rice leafhopper, and yellow stem borer) and diseases (bacterial leaf blight, blast, sheath blight, and tungro). Yield was severely affected in the rice-growing countries of Asia and Africa (Van Oort and Zwart 2018). These biotic stresses significantly cause annual crop losses that threaten global food security. According to the estimation of Roy-Barman and Chattoo (2005), global yield losses annually are equivalent to the quantity required to feed 60 million people. Worldwide, fungal disease alone is estimated at 14% of the annual rice yield decline. Hence, significant changes in GCV lead to increased extreme weather patterns and this combined with increasing air temperatures are the leading causes of the spread of disease into different areas, which has also differed from region to region and from one agroecology to another (Anderson et al. 2004; Agrios 2005). Therefore, exploiting the diverse resources of rice germplasm and understanding the host-plant resistance mechanism at molecular genetics and cellular levels will provide a viable option to manage this disease vis-à-vis the application of various pesticides. For better management of biotic stresses, integrative strategies are required for the selection of resistant rice varieties, and identifying QTLs and genes to understand the genetics of resistance from advanced genotyping technologies and pathogen races. This will deliver valuable information for the development of future climate-smart rice varieties.
Recent progress in QTL and gene/allele identification technologies, marker-assisted selection (MAS), and advanced genomic techniques has been used to develop disease and insect resistance in rice. The molecular genetics of the resistance to the various pathogens has been well documented through conventional or molecular marker-assisted breeding strategies. Therefore, the use of this disease resistance and insect resistance in genomic regions of QTLs and genes provides the most cost-effective and prominent approach for decreasing pesticide use. To date, more than 100 genes for blast resistance, 40 genes for bacterial blight resistance, 34 genes for brown planthopper resistance, 11 genes for general insect resistance, and 6 genes for sheath blight resistance have been identified on different chromosomes (http://qtaro.abr.affrc.go.jp/). Further, some of these genes have been isolated and functionally characterized (http://qtaro.abr.affrc.go.jp/). However, the two major diseases (blast and BLB) frequently affect crop yield and decrease it in most rice-growing regions worldwide. These cloned genes are providing valuable sources to understand the interaction between the disease and host, pathogen mechanism, and, further, as a means to enhance resistance mechanisms in the various backgrounds using marker-assisted breeding and genetic engineering approaches. However, breakdown of the resistance of a single gene has also been identified after 2 or 3 years. This indicates changes in the frequency of pathotypes or the emergence of new ones through mutations and other mechanisms. Therefore, to develop broad-spectrum resistance in rice varieties by stacking multiple resistance genes or QTLs into a single rice variety seeks to provide resistance to a wide range of races as compared to one or two gene combinations.
Pyramiding of multiple resistance genes is a useful molecular breeding strategy for the expression simultaneously of more than one gene to achieve durable resistance against desired target diseases, and it also needs to prevent or delay the breakdown of resistance (Shinada et al. 2014; Feng et al. 2018; Kumar et al. 2018; Liu et al. 2020). Ashkani et al. (2015) have reviewed different successful molecular breeding schemes and gene pyramiding strategies to improve biotic disease resistance genes in rice. Notably, Ji et al. (2016) used a gene pyramiding strategy to develop restorer lines with resistance to multiple diseases : blast (Pita, Pi1, and Pi2), bacterial blight (Xa23 and xa5), and brown planthopper (Bph3). Therefore, restorer lines are useful in hybrid rice breeding programs. In the GSR breeding program, several biotic disease-resistance QTLs and cloned genes have been used in developing novel GSR breeding materials. Pi2, Xa23, Bph14, and Bph15 genes have been mainly introgressed into different GSR lines. Recently, a novel gene, Bph38(t), identified on chromosome 1 explained phenotypic variation of 35.9% in a backcross population derived from a cross of HHZ and Khazar (Balachiranjeevi et al. 2019). The development of molecular mapping and functional genomics of insect resistance revealed that most of the BPH resistance genes were clustered together in specific regions on different chromosomes (3, 4, 6, and 12), except for bph5, bph8, and BPH22(t) to BPH24(t). For example, 8 genes were located in the 19.1–24.4 Mb region on chromosome 12 and 12 genes in the 4.1–8.9 Mb region on chromosome 4 (Du et al. 2020). These clustered genes provide a valuable resource for identifying specific alleles and interaction of the different genes that are involved in the molecular and physiological pathways for the insect resistance mechanism in rice.
4 Molecular Genetics and Breeding Strategies to Combine Multiple Stresses
To develop rice varieties with multiple stress tolerance along with superior grain yield and quality, breeders are exploring different breeding strategies. However, limited progress has been made in this direction through conventional breeding approaches, which are lengthy and laborious processes. Recently available genotyping technologies such as SNP chips, genotyping by sequencing (GBS), and whole-genome sequencing (WGS) have led to the identification of major-effect QTLs associated with the complex abiotic stresses such as drought, salinity, and flooding in rice (Guo et al. 2014; Wang et al. 2017; Ali et al. 2018b; Le Nguyen et al. 2019; Yadav et al. 2019). The identification of molecular markers tightly associated with a trait is valuable for marker-assisted selection. The marker-assisted breeding efforts that began at IRRI led to the identification of a major QTL, Sub1, derived from FR13A, an Indian landrace; qDTY1.1 and qDTY2.1 from drought-tolerant genotype Apo; qDTY12.1 from Way Rarem; and Saltol identified from Pokkali, which showed large effects across different genetic backgrounds. Deployment of these QTLs in molecular breeding strategies paved the way for the development and release of rice varieties such as Swarna-Sub1, Samba Mahsuri-Sub1, Swarna-Sub1, DRR Dhan-50, and CR Dhan 802, which are suitable for different ecosystems (Bhandari et al. 2019). Most of the rice-growing areas in the rainfed environment in South and Southeast Asia are frequently affected by multiple abiotic stresses even within the same cropping season near the coastal areas. Therefore, developing new breeding materials that could tolerate multiple stresses and also provide higher grain yield is essential for global food security.
Over the past decade, there have been only a few significant reports on pyramiding QTLs for stacking multiple traits in the different backgrounds of popular high-yielding rice varieties through MAS approaches. The identified superior and stable lines across different environments and having acceptable grain quality traits are promoted for release in different countries. Pyramiding of the major biotic and abiotic stress-tolerance QTL combinations has been used in different breeding strategies for developing rice varieties with multiple-stress tolerance (Zhu et al. 2015; Dixit et al. 2017; Pang et al. 2017b; Feng et al. 2018; Kumar et al. 2018). Recently, Muthu et al. (2020) developed an improved White Ponni, a popular high-yielding rice variety with significant tolerance against drought, salinity, and submergence by introgressing major-effect QTLs (qDTY1.1, qDTY2.1, Saltol, and Sub1) through a marker-assisted backcross breeding approach. However, the innovative Green Super Rice breeding strategy successfully demonstrated that a high genetic diversity exists within the primary gene pool for improving multiple-stress-tolerance traits, especially for rainfed environments (Ali et al. 2006, 2017). Stringent simultaneous phenotypic selection by screening under multiple abiotic stresses in the early generations has a major advantage in developing trait-specific backcross inbred lines with significantly improved tolerance. This provides an opportunity for the discovery of genes/QTLs underlying the target and non-target traits. With the GSR breeding strategy within the span of 7 years, a total of 27 IRRI-bred GSR varieties were released, and more than 104 rice varieties have been nominated for national cooperative yield trials from three recipient parents and 16 donors. These varieties are now being cultivated on more than 2.7 million ha on a seed distribution basis alone for farmers in Asia and Africa (Ali et al. 2017; Feng et al. 2018; Yu et al. 2020). The integration of advanced genotyping technology such as SNP genotyping array and tGBS in the GSR breeding program provides a high-quality SNP calling accuracy with a low percentage of missing rates across populations. This genotyping information from SNPs is an excellent source for understanding the genetics of green traits and can be used for dissecting many complex traits by using marker-trait association studies (Wing et al. 2018; Feng et al. 2018). Among the various breeding populations, the early backcross-selective introgression lines (EB-SILS) and designed QTL pyramiding (DQP) approach have proven to be an effective strategy for dissecting complex traits such as drought, salinity, and low-temperature stress tolerance; arsenic toxicity tolerance; and nutrient-use efficiency (Dimaano et al. 2017; Pang et al. 2017b; Ali et al. 2018b; Feng et al. 2018; Mahender et al. 2019; Murugaiyan et al. 2019; Najeeb et al. 2020). So far, more than 3200 genes have been listed in Oryzabase, and they were associated with a wide range of stress-tolerance mechanisms in biotic and abiotic stresses (https://shigen.nig.ac.jp/rice/oryzabase/). More than 1800 genes were functionally characterized and deposited in the Q-TARO (QTL Annotation Rice Online) database (http://qtaro.abr.affrc.go.jp/). Of these, cloned rice genes associated with tolerance of multiple biotic stresses (diseases and insect pests) and abiotic stresses (drought, salinity, flooding, anaerobic germination, low nutrient-use efficiency) and also grain quality traits (for eating) are a vital concern in GSR breeding. The availability of rice genome annotation and functionally characterized genes from these two databases are highlighted in Fig. 4. The main focus of green traits involves the genes governing them: 104 for drought tolerance, 95 for salinity tolerance, 60 for low-input tolerance, 52 for cold tolerance, and 8 for submergence tolerance. These are also referred to as green genes, and they mostly represent resource-saving and environment-friendly approaches. As a result of the molecular genetics of green traits in the GSR breeding program, a total of 225 QTLs and 332 candidate genes have been identified on 12 chromosomes, and, trait-wise, each is explained in Table 2 and Fig. 2.
Distribution of cloned genes in rice. (a) The important genes associated with green traits were highlighted across the 12 chromosomes using a Phenogram plot. (b) Traits were associated with the number of genes functionally characterized and these genes provide a valuable resource for understanding the complex nature of stress tolerance and adaptive traits. (Source: https://orygenesdb.cirad.fr/data.html and http://qtaro.abr.affrc.go.jp/)
In the genomic region of these QTL positions, a large number of candidate genes were identified through the in silico database and many of them have known functions related to green traits. Also, a few of them have unknown functions. The comprehensive literature survey and rice genomic database showed that genes such as OsCOIN, OsDREB2A, OsGSK1, and OsDREB1F on chromosome 1; OsGS1;2, OsMYB2, and ZFP182 on chromosome 3; OsTPS1 on chromosome 5; OsDREB1C and OsiSAP8 on chromosome 6; OsDREB1B and OsDREB1A on chromosome 9; and OsNAC5 on chromosome 11 are responsible for the multiple stress-tolerance mechanism. These genes act in ABA signaling pathways, hormonal regulation, accumulation of sugar and other compatible solutes such as proline, and also many other developmental and physiological processes involved in the regulation of the multiple stress-tolerance mechanism. Most of the multiple stress-tolerance gene expression is induced by ABA, and it depends on the presence of a cis-acting element referred to as ABA-responsive element (ABRE). These ABA-independent and dependent signaling pathways are involved in stress responses including drought, cold, heat, and cold. However, certain genomic regions on chromosome 1 (33.16–33.97 Mb; 40.15–41.90 Mb), chromosome 3 (11.07–11.75 Mb; 35.01–35.56 Mb), and chromosome 9 (21.13–21.98 Mb) played a significant role in the biotic and abiotic stress-tolerance pathways. They mainly played a role in major ABA signaling pathways and other stress signal transduction mechanisms in regulating stress tolerance and crosstalk between the other transcription factors to enhance gene regulation against multiple stresses. These overlapping stress-tolerance genes and transcription factors regulating similar stress-tolerance signaling pathways exist besides the crosstalk among the biotic and abiotic stresses. However, the hotspot genomic regions of cloned genes and their validation with earlier reports will provide an improved understanding of the molecular and physiological mechanisms in response to stress tolerance to help in the improvement of grain yield and quality traits in rice.
4.1 Dissecting the Stress-Regulated Mechanisms for Multiple Stress Tolerance
Simultaneous exposure to a single or multiple biotic and abiotic stresses strongly affects crop production, mainly in the rice-growing countries of Asia and Africa. In most of the rice-growing regions in the rainfed environment, drought stress is a major factor affecting about 23 million hectares in Southeast Asia and, combined with biotic stresses such as diseases and insects, along with rising temperatures, could further significantly decrease grain yield (Aghamolki et al. 2014; Bahuguna et al. 2018). The response of the plant’s tolerance mechanism and adaptive strategies toward multiple stresses are significantly limited. It is crucial to understand this at the genomics and metabolic levels because of non-additive interactions, extensive overlaps, and crosstalk between stress-response signaling pathways, and also the interactions between transcription factors (TFs) and cis-elements on the promoters of target genes (Kissoudis et al. 2014; Verma and Deepti 2016). Generally, the combination of biotic and abiotic stress impacts depends on two factors: the host tolerance/susceptibility mechanism and the interaction of plant-microbial reactions, which influence stress responses to plants. However, a few common morpho-physiological traits such as leaf wilting, tiller number, harvest index, root growth pattern, and chlorophyll content have been identified in the case of both drought and bacterial infections. They further decrease the photosynthetic machinery and diminish grain yield (Pandey et al. 2017). Importantly, the central role of hormonal balance and interaction is the critical controller of genes that play a significant role in regulating the stress-tolerance genes that are involved in different molecular and physiological mechanisms for multiple stresses. Hormonal interaction is regulated by ethylene, ABA, salicylic acid, jasmonic acid, cytokinin, and brassinosteroid. They, in turn, regulate several growth development traits. They also connect to the multiple stress signaling pathways to regulate stress-responsive gene expression (Vemanna et al. 2019). The receptor of these hormones regulates the various TF families such as NAC, AP2/ERF, bZIP, and MYC, which have altered the stress response to biotic and abiotic stresses (Huang et al. 2011; Li et al. 2018). However, integration of the omics approach has provided more profound insights into the molecular mechanisms for a better understanding of the multiple stress-responsive candidate genes. Further, this helped in the functional characterization of each gene that is involved in the stress signaling and tolerance mechanisms for abiotic and biotic stress adaptation.
Recently, Vemanna et al. (2019) reviewed comprehensive information on crosstalk signaling and multiple stress-tolerance mechanisms in rice. The overlapping stress-tolerance mechanisms and transcriptional responses in combined stresses are quite complex and interact with several biological processes. There are few reports on a meta-analysis using transcriptome data, and they found certain common genomic regions shared with multiple or individual stress tolerances. This indicates that, in response to stress tolerance, several hormonal signaling pathways are overlapping and cross-talking. Interestingly, Zhang et al. (2016) identified a total of 178 genes that were commonly expressed in drought and bacterial pathogen infections from the transcriptome analysis. However, some genes play specific functional and opposite roles in biotic and abiotic stresses . For example, one of the CDPK family proteins, OsCPK12, regulates drought, salinity, and cold stress tolerance, but it is a negative regulator for blast resistance (Asano et al. 2012; Fang et al. 2019). Similarly, the WRKY transcription factor family of WRKY71 is responsible for increasing the tolerance of bacterial infections, whereas the overexpression of WRKY45 showed susceptibility. Universal stress-tolerance proteins such as ABC transporters play a vital role in the development of stress tolerance in both biotic and abiotic stresses.
In the GSR breeding strategy, Murugaiyan et al. (2019) identified a robust QTL for arsenic tolerance on chromosome 1 and found a multi-drug resistance-associated protein (OsMRP2), which belongs to the subfamily of ABC transporters. It is mainly involved in the vacuolar sequestration of toxic metabolites (Brunetti et al. 2015). In the GSR breeding program, we primarily focused on dissecting complex abiotic traits. The rice crop is susceptible to cold stress, which adversely affects the crop at various growth stages, causing significant yield decreases mainly in temperate, tropical, and subtropical rice-growing regions. Similary, Najeeb et al. (2020) identified low-temperature stress-tolerance QTLs and found some promising genomic regions that were involved in the multiple stress-tolerance mechanism in rice. For instance, the major QTL on chromosome 5 had a possible candidate gene, Os05g49970, encoding translation initiation factor-2 (eIF2). These genes are involved in several cellular and metabolic processes in the early growth developmental stages, hormonal signaling pathways in plant defense mechanisms, and tolerance of various abiotic stresses (Martínez-Silva et al. 2012; Mutuku et al. 2015). Likewise, on chromosome 6, two genes (Os06g17220 and Os06g48300) were involved in major ABA-dependent signaling pathways and were responsible for sucrose-synthetic activity during anoxia conditions (Lasanthi-Kudahettige et al. 2007; Bhatnagar et al. 2017). Zhang et al. (2011) had attempted to integrate contemporary knowledge of signal transduction pathways with the principles of quantitative and population genetics to illustrate the genetic networks underlying complex traits using a model established upon the one-way functional dependency of downstream genes on upstream regulators depicting the principle of hierarchy. The mutual functional dependence among related genes was determined as the functional genetic units (FGUs). Interestingly, both simulated and real data suggested that complementary epistasis contributes significantly to quantitative trait variation and obscures the phenotypic effects of many “downstream” loci in pathways. Downstream FGUs were more vulnerable to loss of function than their upstream regulators; however, this vulnerability was compensated by different FGUs of similar functions (Zhang et al. 2011). Dissecting the complex trait of nutrient-use efficiency under low-input conditions (Mahender et al. 2019) identified a key regulator as F-box protein and calcium-dependent protein kinase on chromosome 2. These genomic regions played a major role in carbon and nitrogen metabolism and also maintained the homeostatic coordination of other enzymatic activities. The integrated approaches of omics and gene network analysis could play a crucial role in understanding stress tolerance in combined or individual stress-tolerance mechanisms in rice. These strategies need to be assessed more to gain deeper insights into dissecting the complexity and expanding the knowledge on each specific role of stress-responsive genes and transcription factors. Placing all the outputs from these strategies together could help to understand the unique and shared molecular and physiological pathways and possibly increase the adaptation to multiple abiotic and biotic stress factors.
4.2 Breeding Products Combining Tolerance of Multiple Stresses
Maintaining genetic diversity in breeding programs is a critical component, and it provides an excellent opportunity for breeders to develop novel and improved cultivars with desirable target traits in their breeding program. The GSR breeding program contains two interlinked breeding strategies: selective introgression lines (SILs) and the designed QTL pyramiding (DQP) strategy. First is to develop multi-trait-specific SILs from several BC1F2 bulk populations derived from a few highly adapted recipient parents and 10–15 donor parents with three rounds of multiple-stress screening. This results in the development of EB-SILs, and these high-yielding, multi-stress-tolerant BC1F5 SILs then undergo two seasons of preliminary yield trials and advanced yield trials to identify superior lines simultaneously across drought, low-input, and irrigated conditions. The superior SILs that outyield the standard checks under different conditions are shortlisted for global multi-location trials. Superior multi-trait-specific SILs are crossed with another IL with complementary traits from within the same recipient/donor combination or different donors. Such a designed cross based on genotypic and phenotypic information is referred to as a DQP approach. This has played a vital role in increasing grain yield, improving tolerance of abiotic and biotic stresses, and using fewer inputs such as fertilizer and pesticide. This breeding strategy began in 2008 at IRRI and was used to create breeding materials that involved 500 elite rice varieties that belong to the mini-core collection and are part of the 3000 Rice Genomes Project (Ali et al. 2012). Among these, only three recipient parents, Huanghuazhan (HHZ), Weed Tolerant Rice-1 (WTR-1), and TME80518, and 16 donors were fully used for the EBBP and DQP approach to identify promising GSR materials with multiple abiotic and biotic stress tolerance (Ali et al. 2012, 2013b, 2018a) (Fig. 3). The BC1F2 populations underwent three rounds of precise phenotypic screening under drought, salinity, submergence, low-chemical-input, and normal irrigated conditions. This led to identifying 845 trait-specific SILs that outperformed the tolerant checks and these were further evaluated under multi-environment locations in Asian and African countries. As of now, a total of 66 GSR breeding materials have been registered in China and 59 GSR breeding materials have been released across Asian and African countries (Fig. 5). More than 90 GSR rice varieties were nominated in national cooperative yield trials from 2016 to 2018. The released varieties showed consistency in higher grain yield under low inputs (fertilizer and pesticide) and tolerance of multiple biotic and abiotic stresses. For instance, Yorobe et al. (2016) assessed the impact of GSR varieties and evaluated the income of farmer-users per hectare. Based on the survey data and fixed-effect model approach, farmer income increased significantly compared with that of farmers using conventional inbred varieties (non-GSR materials) with the frequent occurrence of flood and submergence conditions (Yorobe et al. 2014; Kodama et al. 2019). In addition, the net farm income advantage with GSR is quite high under increasing percentiles of rainfall. Several GSR varieties outperformed the local checks (Table 2). One GSR line (IRIS 179-880151) in data from two seasons for 2 years showed a 10% yield advantage vis-à-vis the local check variety and also had tolerance of drought, salinity, and submergence (Ali et al. 2013a). Similarly, multiple-stress-tolerant lines such as GSR IR1-12-D10-S1-D1, GSR IR1-5-S10-D1-D1, and GSR IR1-8-S12-Y2-D1 performed well under different environments and had a higher grain yield advantage of 25% to 40% over the drought-tolerant checks (Marcaida III et al. 2014). Another recent example of the potential impact of GSR varietal performance is found in the sub-Saharan African region of Mozambique, which is one of the major rice exporters. Kodama (2019) conducted a farm-level survey in three regions (Gaza, Sofala, and Nampula) and used an endogenous switching regression (ESR) model to assess GSR varieties among smallholder rice farmers and smallholder rice producers. Interestingly, smallholders carry out about 90% of the rice production in this region, and average yield is 1.0–1.2 t/ha in rainfed systems and 2.8–3.5 t/ha in normal irrigated conditions. This region is more prone to abiotic stresses, and mainly to the variability and duration of rainfall. With the adoption of GSR varieties, Kodama (2019) noticed a significant yield advantage, about ten times higher for smallholder farmers than for smallholder rice producers adopting non-GSR varieties. This indicates the strong and positive yield advantage and tolerance of multiple stresses across Asian and African countries that benefit poor farmers’ income. This provides a great opportunity for GSR varieties to provide more rice and help alleviate poverty across the major rice-growing countries.
Achievements of GSR breeding program and released GSR varieties across the globe. (a) Map showing the number of released GSR varieties highlighted country-wise. (b) In a span of 6 years, the total number of GSR varieties released and nominated in national cooperative yield trials across Asia and Africa
4.3 Development of Rice Hybrids with Multiple-Stress Tolerance
Hybrid rice breeding is one of the major core components for ensuring global food security. From a historical perspective, two milestones have been reached in enhancing grain yield. The first was by developing semi-dwarf rice varieties through the incorporation of the semi-dwarf (sd1) gene in rice breeding at IRRI in the early 1960s and the second was by the use of heterosis in hybrid rice (Yuan 2017). The potential of these two strategies was shown by rice grain yield increasing by about 30% with the semi-dwarf gene and a 15–20% yield increase with the use of heterosis in the major rice-producing areas of China (Peng et al. 2009). Currently, there are three approaches for increasing hybrid vigor: the cytoplasmic male sterility/fertility restoration (CMS/Rf) locus, environment-sensitive genic male sterility, and apomixis (Birchler et al. 2006; Bar-Zvi et al. 2017; Xie et al. 2019). At IRRI, we began developing climate-smart rice hybrids by adopting EB-SILs and using the DQP approach in separate restorer and maintainer backgrounds (Ali et al. 2020). The promising rice varieties that are tolerant of multiple stresses and possess higher grain yield and superior grain quality traits were considered for parental selection based on the fertility restoration locus and possession of desirable floral characteristics. Based on genotyping results, the GSR lines positive for fertility restoration loci Rf3 and Rf4 will be considered as restorers and those entirely negative for those loci as maintainers. More than 100 restorer lines with a strong general combining ability, tolerance of multiple stresses, and having preferable grain quality have been bred from using the promising lines from the GSR breeding program. These climate-smart parental lines are essential for complementing the introgression of QTLs and genes with green traits into the F1s. Hybrid rice-related traits governing wide compatibility, floral traits that promote outcrossing, and seed reproducibility are important for parental line-breeding besides addressing the market requirements of the target regions. Further, by selecting the maximum genetic distance between the parental lines from diverse heterotic pools, this helped to identify promising high-yielding climate-resilient hybrid rice combinations. Recently, genomic hybrid breeding has emerged using whole-genome markers to predict future hybrids. Only the superior predicted hybrids are then field-evaluated and later released as new hybrid cultivars based on their actual performance in the target conditions. This approach offers an opportunity to select truly superior hybrids at limited cost. Cui et al. (2019) used genomic best linear unbiased prediction to predict hybrid performance using an existing rice population of 1495 hybrids. Further, a replicated tenfold cross-validation showed that the prediction ability on ten agronomic traits ranged from 0.35 to 0.92 (Cui et al. 2019). By keeping this population of 1495 hybrids, it can be used to predict hybrids from seemingly unrelated parents. Machine learning algorithms and artificial intelligence are also being employed for predicting the best hybrid combinations at IRRI using genomic and historical hybrid yield trial data.
5 Conclusions
Identifying the promising genomic regions of QTLs and genes that are associated with the green traits that are involved in multiple-stress tolerance mechanisms will be adapted to increase rice yield in the future. The Green Super Rice breeding strategy using EB-SILs and DQP, along with combined advanced genomic technologies, significantly improved the development of rice varieties with multiple-stress tolerance. This breeding strategy also helped to identify promising QTLs and genes that were associated with the green traits related to higher grain yield, tolerance of abiotic and biotic stresses, low-input use of fertilizer and pesticide, and superior grain quality. These resource-saving and environment-friendly features have been well exploited under the GSR breeding strategy. The stress-tolerance mechanism of these combined or individual stresses that triggers a network of signaling pathways at the plant cell level needs to be studied in detail. The crosstalk of these biotic and abiotic stresses could reveal similar pathways for maintaining the cellular mechanism for tolerance. Omics approaches should occupy the core component in breeding and trait development activities for identifying multiple-stress tolerance, and this would remain the primary target trait in the next generation of crop breeding programs.
References
Aghamolki MTK, Yusop MK, Oad FC et al (2014) Heat stress effects on yield parameters of selected rice cultivars at reproductive growth stages. J Food Agric Environ 12:741–746
Agrios GN (2005) Plant pathology, 5th edn. Elsevier Academic Press, Burlington, pp 79–103
Ali AJ, Xu JL, Ismail AM et al (2006) Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Res 97:66–76
Ali J, Xu JL, Gao YM et al (2012) Green super rice (GSR) technology: an innovative breeding strategy-achievements & advances. In: The 12th SABRAO congress on plant breeding towards 2025: challenges in a rapidly changing world. Chiang Mai, Thailand. pp 16–17
Ali J, Xu JL, Gao YM et al (2013a) Breeding for yield potential and enhanced productivity across different rice ecologies through green super rice (GSR) breeding strategy. In: Muralidharan K, Siddiq EA (eds) International dialogue on perception and prospects of designer rice. Society for Advancement of Rice Research, Hyderabad, pp 60–68
Ali N, Paul S, Gayen D et al (2013b) RNAi mediated down regulation of myo-inositol-3-phosphate synthase to generate low phytate rice. Rice 6:12. https://doi.org/10.1186/1939-8433-6-12
Ali J, Xu J-L, Gao Y-M et al (2017) Harnessing the hidden genetic diversity for improving multiple abiotic stress tolerance in rice (Oryza sativa L.). PLoS One 12:e0172515. https://doi.org/10.1371/journal.pone.0172515
Ali J, Aslam UM, Tariq R et al (2018a) Exploiting the genomic diversity of rice (Oryza sativa L.): SNP-typing in 11 early-backcross introgression-breeding populations. Front Plant Sci 9:1–10. https://doi.org/10.3389/fpls.2018.00849
Ali J, Jewel ZA, Mahender A et al (2018b) Molecular genetics and breeding for nutrient use efficiency in rice. Int J Mol Sci 19:1762. https://doi.org/10.3390/ijms19061762
Ali J, Mahender A, Prahalada GD et al (2020) Genomics-assisted breeding of climate-smart inbred and hybrid rice varieties. In: Kole C (ed) Genomic designing of climate-smart cereal crops. Springer, Cham, Springer Nature Switzerland AG, pp 1–43. https://doi.org/10.1007/978-3-319-93381-8_1
Anderson PK, Cunningham AA, Patel NG et al (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol 19:535–544
Antralina M, Istina IN, Simarmata T (2015) Effect of difference weed control methods to yield of lowland rice in the SOBARI. Procedia Food Sci 3:323–329
Appleby AP, Müller F, Carpy S (2002) Weed control. Ullmann’s encyclopedia of industrial chemistry. Wiley, Weinheim
Asano T, Hayashi N, Kobayashi M et al (2012) A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J 69:26–36
Ashkani S, Rafii MY, Shabanimofrad M et al (2015) Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front Plant Sci 6:886
Bahuguna RN, Gupta P, Bagri J et al (2018) Forward and reverse genetics approaches for combined stress tolerance in rice. Indian J Plant Physiol 23:630–646
Bailey-Serres J, Fukao T, Ronald P et al (2010) Submergence tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice 3:138–147
Balachiranjeevi CH, Prahalada GD, Mahender A et al (2019) Identification of a novel locus, BPH38 (t), conferring resistance to brown planthopper (Nilaparvata lugens Stal.) using early backcross population in rice (Oryza sativa L.). Euphytica 215:185
Balakrishnan D, Subrahmanyam D, Badri J et al (2016) Genotype × environment interactions of yield traits in backcross introgression lines derived from Oryza sativa cv. Swarna/Oryza nivara. Front Plant Sci 7:1–19. https://doi.org/10.3389/fpls.2016.01530
Balyan HS, Gupta PK, Kumar S et al (2013) Genetic improvement of grain protein content and other health-related constituents of wheat grain. Plant Breed 132:446–457
Bar-Zvi D, Lupo O, Levy AA, Barkai N (2017) Hybrid vigor: the best of both parents, or a genomic clash? Curr Opin Syst Biol 6:22–27
Bazrkar-Khatibani L, Fakheri BA, Hosseini-Chaleshtori M et al (2019) Genetic mapping and validation of quantitative trait loci (QTL) for the grain appearance and quality traits in rice (Oryza sativa L.) by using recombinant inbred line (RIL) population. Int J Genomics 2019:3160275. https://doi.org/10.1155/2019/3160275
Bernier J, Kumar A, Ramaiah V et al (2007) A large effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci 47:507–516. https://doi.org/10.2135/cropsci2006.07.0495
Bhandari A, Jayaswal P, Yadav N et al (2019) Genomics-assisted backcross breeding for infusing climate resilience in high-yielding green revolution varieties of rice. Indian J Genet 79(1 Suppl):160–170
Bhatnagar N, Min M-K, Choi E-H et al (2017) The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol Biol 93:389–401
Birchler JA, Yao H, Chudalayandi S (2006) Unraveling the genetic basis of hybrid vigor. Proc Natl Acad Sci U S A 103:12957–12958
Brunetti P, Zanella L, De Paolis A et al (2015) Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J Exp Bot 66(13):3815–3829. https://doi.org/10.1093/jxb/erv185
Calayugan MIC, Formantes AK, Amparado A et al (2020) Genetic analysis of agronomic traits and grain iron and zinc concentrations in a doubled haploid population of rice (Oryza sativa L.). Sci Rep 10:1–14
Chauhan BS, Abugho SB (2013) Weed management in mechanized-sown, zero-till dry-seeded rice. Weed Technol 27:28–33
Chauhan BS, Awan TH, Abugho SB, Evengelista G (2015a) Effect of crop establishment methods and weed control treatments on weed management, and rice yield. Field Crops Res 172:72–84
Chauhan BS, Opeña J, Ali J et al (2015b) Response of 10 elite “Green Super Rice” genotypes to weed infestation in aerobic rice systems. Plant Prod Sci 18:228–233. https://doi.org/10.1626/pps.18.228
Chen S, Yang Y, Shi W et al (2008) Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell 20(7):1850–1861
Chen X, Cui Z, Fan M et al (2014a) Producing more grain with lower environmental costs. Nature 514:486–489
Chen L, Lin L, Cai G et al (2014b) Identification of nitrogen, phosphorus, and potassium deficiencies in rice based on static scanning technology and hierarchical identification method. PLoS One 9:e113200. https://doi.org/10.1371/journal.pone.0113200
Cheng SH, Zhuang JY, Fan YY et al (2007) Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot 100:959–966
Cohen SP, Leach JE (2019) Abiotic and biotic stresses induce a core transcriptome response in rice. Sci Rep 9:1–11
Cuevas RP, Pede VO, McKinley J et al (2016) Rice grain quality and consumer preferences: a case study of two rural towns in the Philippines. PLoS One 11:e0150345
Cui K, Peng S, Xing Y et al (2002) Molecular dissection of seedling-vigor and associated physiological traits in rice. Theor Appl Genet 105:745–753
Cui Y, Li R, Li G et al (2019) Hybrid breeding of rice via genomic selection. Plant Biotechnol J 18:57–67
Custodio MC, Cuevas RP, Ynion J et al (2019) Rice quality: how is it defined by consumers, industry, food scientists, and geneticists? Trends Food Sci Technol 92:122–137
da Cruz RP, Sperotto RA, Cargnelutti D et al (2013) Avoiding damage and achieving cold tolerance in rice plants. Food Energy Secur 2:96–119. https://doi.org/10.1002/fes3.25
Dang X, Thi TGT, Dong G et al (2014) Genetic diversity and association mapping of seed vigor in rice (Oryza sativa L.). Planta 239:1309–1319
Dimaano NGB, Ali J, Cruz PCSS et al (2017) Performance of newly developed weed-competitive rice cultivars under lowland and upland weedy conditions. Weed Sci 65:798–817. https://doi.org/10.1017/wsc.2017.57
Dimaano NGB, Ali J, Mahender A et al (2020) Identification of quantitative trait loci governing early germination and seedling vigor traits related to weed competitive ability in rice. Euphytica 216:159. https://doi.org/10.1007/s10681-020-02694-8
Diwan J, Channbyregowda M, Shenoy V et al (2013) Molecular mapping of early vigour related QTLs in rice. Res J Biol 1:24–30
Dixit S, Singh A, Cruz MTS et al (2014) Multiple major QTL lead to stable yield performance of rice cultivars across varying drought intensities. BMC Genet 15:16
Dixit S, Singh A, Sandhu N et al (2017) Combining drought and submergence tolerance in rice: marker-assisted breeding and QTL combination effects. Mol Breed 37:143
Du B, Chen R, Guo J, He G (2020) Current understanding of the genomic, genetic, and molecular control of insect resistance in rice. Mol Breed 40:24
Elferink M, Schierhorn F (2016) Global demand for food is rising. Can we meet it? Harv Bus Rev 7:2016
Ella ES, Kawano N, Osamu H (2003) Importance of active oxygen scavenging system in the recovery of rice seedlings after submergence. Plant Sci 165:85–93
Fahad S, Bajwa AA, Nazir U et al (2017) Crop production under drought and heat stress: plant responses and management options. Front Plant Sci 8:1147
Fahad S, Adnan M, Noor M et al (2019) Major constraints for global rice production. In: Advances in rice research for abiotic stress tolerance. Elsevier, Amsterdam, pp 1–22
Fang WB, Yong CL, Hua CS et al (2019) Impaired function of the calcium-dependent protein kinase, OsCPK12, leads to early senescence in rice (Oryza sativa L.). Front Plant Sci 10:52
Farooq M, Siddique KHM, Rehman H et al (2011) Rice direct seeding: experiences, challenges and opportunities. Soil Tillage Res 111:87–98
Feng B, Chen K, Cui Y et al (2018) Genetic dissection and simultaneous improvement of drought and low nitrogen tolerances by designed QTL pyramiding in rice. Front Plant Sci 9:306. https://doi.org/10.3389/fpls.2018.00306
Fischer RA, Edmeades GO (2010) Breeding and cereal yield progress. Crop Sci 50:S-85
Foolad MR, Subbiah P, Zhang L (2007) Common QTL affect the rate of tomato seed germination under different stress and nonstress conditions. Int J Plant Genomics 2007:97386. https://doi.org/10.1155/2007/97386
Gao Z, Zeng D, Cheng F et al (2011) ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J Integr Plant Biol 53:756–765
Gautam T, Saripalli G, Kumar A et al (2020) Introgression of a drought insensitive grain yield QTL for improvement of four Indian bread wheat cultivars using marker assisted breeding without background selection. J Plant Biochem Biotechnol 29:1–12. https://doi.org/10.1007/s13562-020-00553-0
Ghimire KH, Quiatchon LA, Vikram P et al (2012) Identification and mapping of a QTL (qDTY1.1) with a consistent effect on grain yield under drought. Field Crops Res 131:88–96
Gimhani DR, Gregorio GB, Kottearachchi NS, Samarasinghe WLG (2016) SNP-based discovery of salinity-tolerant QTLs in a bi-parental population of rice (Oryza sativa). Mol Gen Genomics 291:2081–2099
Gothandam KM, Kim E-S, Cho H, Chung Y-Y (2005) OsPPR1, a pentatricopeptide repeat protein of rice, is essential for the chloroplast biogenesis. Plant Mol Biol 58:421–433
Guan YS, Serraj R, Liu SH et al (2010) Simultaneously improving yield under drought stress and non-stress conditions: a case study of rice (Oryza sativa L.). J Exp Bot 61:4145–4156. https://doi.org/10.1093/jxb/erq212
Guo L, Gao Z, Qian Q (2014) Application of resequencing to rice genomics, functional genomics and evolutionary analysis. Rice 7:4. https://doi.org/10.1186/s12284-014-0004-7
Guo J, Hu X, Gao L et al (2017) The rice production practices of high yield and high nitrogen use efficiency in Jiangsu, China. Sci Rep 7:2101
Guojun P, Shuqiang C, Chengyan S et al (2009) Study on relationship between resistance to blast and yield traits in early japonica rice in cold region [J]. Chin Agric Sci Bull 19
Hawkesford MJ, Griffiths S (2019) Exploiting genetic variation in nitrogen use efficiency for cereal crop improvement. Curr Opin Plant Biol 49:35–42
Huang XS, Luo T, Fu XZ et al (2011) Cloning and molecular characterization of a mitogen-activated protein kinase gene from Poncirus trifoliata whose ectopic expression confers dehydration/drought tolerance in transgenic tobacco. J Exp Bot 62:5191–5206. https://doi.org/10.1093/jxb/err229
Jabran K, Chauhan BS (2015) Weed management in aerobic rice systems. Crop Prot 78:151–163
Jena KK, Kim SM, Suh JP, Kim YG (2010) Development of cold-tolerant breeding lines using QTL analysis in rice. Second Africa Rice Congr 22–26
Jewel Z, Ali J, Pang Y et al (2018) Developing Green Super Rice varieties with high nutrient use efficiency by phenotypic selection under varied nutrient conditions. Crop J 7(3):368–377. https://doi.org/10.20944/preprints201807.0216.v1
Jewel ZA, Ali J, Mahender A et al (2019) Identification of quantitative trait loci associated with nutrient use efficiency traits, using SNP markers in an early backcross population of rice (Oryza sativa L.). Int J Mol Sci 20:900. https://doi.org/10.3390/ijms20040900
Ji Z, Yang S, Zeng Y et al (2016) Pyramiding blast, bacterial blight and brown planthopper resistance genes in rice restorer lines. J Integr Agric 15:1432–1440
Kavurikalpana TS, Shashidhara N (2018) Validation of molecular markers linked to grain quality traits in rice (Oryza sativa L.). Int J Curr Microbiol Appl Sci 7:1897–1902
Khan FA, Narayan S, Bhat SA, Maqbool R (2016) Vermipriming: a noble technology for seed invigouration in rice (Oryza sativa L.). SKUAST J Res 18:124–129
Kissoudis C, van de Wiel C, Visser RGF, van der Linden G (2014) Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front Plant Sci 5:207
Kodama W, Pede VO, Mishra AK, Cabrera ER (2019) Assessing the benefits of Green Super Rice in Sub-Saharan Africa: evidence from Mozambique. Selected paper prepared for presentation at the 2019 Agricultural & Applied Economics Association Annual Meeting, Atlanta, Georgia, July 21–23. 23 p
Kreuzwieser J, Gessler A (2010) Global climate change and tree nutrition: influence of water availability. Tree Physiol 30:1221–1234
Kumar A, Dixit S, Ram T et al (2014) Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J Exp Bot 65:6265–6278. https://doi.org/10.1093/jxb/eru363
Kumar A, Sandhu N, Yadav S et al (2017a) Rice varietal development to meet future challenges. In: The future rice strategy for India. Elsevier, Amsterdam, pp 161–220
Kumar M, Gho Y-S, Jung K-H, Kim S-R (2017b) Genome-wide identification and analysis of genes, conserved between japonica and indica rice cultivars, that respond to low-temperature stress at the vegetative growth stage. Front Plant Sci 8:1120
Kumar A, Sandhu N, Dixit S et al (2018) Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice (N Y) 11:35
Kumar A, Sandhu N, Venkateshwarlu C et al (2020) Development of introgression lines in high yielding, semi-dwarf genetic backgrounds to enable improvement of modern rice varieties for tolerance to multiple abiotic stresses free from undesirable linkage drag. Sci Rep 10:1–13
Lafitte HR, Li ZK, Vijayakumar CHM et al (2006) Improvement of rice drought tolerance through backcross breeding: evaluation of donors and selection in drought nurseries. Field Crops Res 97:77–86. https://doi.org/10.1016/j.fcr.2005.08.017
Lasanthi-Kudahettige R, Magneschi L, Loreti E et al (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144:218–231
Le Nguyen K, Grondin A, Courtois B, Gantet P (2019) Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci 24:263–274
Li Z (1998) Molecular analysis of epistasis affecting complex traits. In: Molecular dissection of complex traits. CRC Press, Boca Raton, pp 119–130
Li Z, Ali J (2017) Breeding green super rice (GSR) varieties for sustainable rice cultivation. In: Sasaki T (ed) Achieving sustainable cultivation of rice, vol 1 edn. Burleigh Dodds Science Publishing, Cambridge, pp 131–152
Li ZK, Fu BY, Gao YM et al (2005) Genome-wide introgression lines and their use in genetic and molecular dissection of complex phenotypes in rice (Oryza sativa L.). Plant Mol Biol 59:33–52. https://doi.org/10.1007/s11103-005-8519-3
Li S, Tian Y, Wu K et al (2018) Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560:595. https://doi.org/10.1038/s41586-018-0415-5
Liang Y, Meng L, Lin X et al (2018) QTL and QTL networks for cold tolerance at the reproductive stage detected using selective introgression in rice. PLoS One 13:e0200846
Liu C, Ding S, Zhang A et al (2020) Development of nutritious rice with high zinc/selenium and low cadmium in grains through QTL pyramiding. J Integr Plant Biol 62:349–359
Mahender A, Anandan A, Pradhan SK (2015) Early seedling vigour, an imperative trait for direct-seeded rice: an overview on physio-morphological parameters and molecular markers. Planta 241:1027–1050
Mahender A, Anandan A, Pradhan SK, Pandit E (2016) Rice grain nutritional traits and their enhancement using relevant genes and QTLs through advanced approaches. Springerplus 5(1):2086. https://doi.org/10.1186/s40064-016-3744-6
Mahender A, Ali J, Prahalada GD et al (2019) Genetic dissection of developmental responses of agro-morphological traits under different doses of nutrient fertilizers using high-density SNP markers. PLoS One 14(7):e0220066
Marcaida M III, Li T, Angeles O et al (2014) Biomass accumulation and partitioning of newly developed Green Super Rice (GSR) cultivars under drought stress during the reproductive stage. Field Crops Res 162:30–38. https://doi.org/10.1016/j.fcr.2014.03.013
Martínez-Silva AV, Aguirre-Martínez C, Flores-Tinoco CE et al (2012) Translation initiation factor AteIF(iso)4E is involved in selective mRNA translation in Arabidopsis thaliana seedlings. PLoS One 7(2):e31606. https://doi.org/10.1371/journal.pone.0031606
Matsushima K-I, Sakagami J-I (2013) Effects of seed hydropriming on germination and seedling vigor during emergence of rice under different soil moisture conditions. Am J Plant Sci 4:1584. https://doi.org/10.4236/ajps.2013.48191
Matthus E, Wu LB, Ueda Y et al (2015) Loci, genes, and mechanisms associated with tolerance to ferrous iron toxicity in rice (Oryza sativa L.). Theor Appl Genet 128:2085–2098. https://doi.org/10.1007/s00122-015-2569-y
Mehta S, Singh B, Dhakate P et al (2019) Rice, marker-assisted breeding, and disease resistance. In: Wani SH (ed) Disease resistance in crop plants. Springer, Cham, pp 83–111
Mogga M, Sibiya J, Shimelis H et al (2018) Diversity analysis and genome-wide association studies of grain shape and eating quality traits in rice (Oryza sativa L.) using DArT markers. PLoS One 13:e0198012
Molla KA, Debnath AB, Ganie SA, Mondal TK (2015) Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice (Oryza sativa L.). BMC Plant Biol 15:122
Mukamuhirwa A, Persson Hovmalm H, Bolinsson H et al (2019) Concurrent drought and temperature stress in rice—a possible result of the predicted climate change: effects on yield attributes, eating characteristics, and health promoting compounds. Int J Environ Res Public Health 16:1043
Murugaiyan V, Ali J, Mahender A et al (2019) Mapping of genomic regions associated with arsenic toxicity stress in a backcross breeding populations of rice (Oryza sativa L.). Rice 12:61
Muthu V, Abbai R, Nallathambi J et al (2020) Pyramiding QTLs controlling tolerance against drought, salinity, and submergence in rice through marker assisted breeding. PLoS One 15:e0227421
Mutuku JM, Yoshida S, Shimizu T et al (2015) The WRKY45-dependent signaling pathway is required for resistance against Striga hermonthica parasitism. Plant Physiol 168:1152–1163
Najeeb S, Ali J, Mahender A et al (2020) Identification of main-effect quantitative trait loci (QTLs) for low-temperature stress tolerance germination- and early seedling vigor-related traits in rice (Oryza sativa L.). Mol Breed 40:10
Nandi S, Subudhi PK, Senadhira D et al (1997) Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol Gen Genet 255(1):1–8
Oladosu Y, Rafii MY, Samuel C et al (2019) Drought resistance in rice from conventional to molecular breeding: a review. Int J Mol Sci 20:3519. https://doi.org/10.3390/ijms20143519
Oliver SN, Dennis ES, Dolferus R (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol 48:1319–1330
Ouk M, Basnayake J, Tsubo M et al (2006) Use of drought response index for identification of drought tolerant genotypes in rainfed lowland rice. Field Crops Res 99:48–58
Panda BB, Sharma S, Mohapatra PK, Das A (2012) Application of excess nitrogen, phosphorus, and potassium fertilizers leads to lowering of grain iron content in high-yielding tropical rice. Commun Soil Sci Plant Anal 43:2590–2602
Pandey P, Irulappan V, Bagavathiannan MV, Senthil-Kumar M (2017) Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci 8:537
Pang Y, Ali J, Wang X et al (2016) Relationship of rice grain amylose, gelatinization temperature and pasting properties for breeding better eating and cooking quality of rice varieties. PLoS One 11:e0168483
Pang Y, Chen K, Wang X et al (2017a) Recurrent selection breeding by dominant male sterility for multiple abiotic stresses tolerant rice cultivars. Euphytica 213:268. https://doi.org/10.1007/s10681-017-2055-5
Pang Y, Chen K, Wang X et al (2017b) Simultaneous improvement and genetic dissection of salt tolerance of rice (Oryza sativa L.) by designed QTL pyramiding. Front Plant Sci 8:1275. https://doi.org/10.3389/fpls.2017.01275
Peng S, Tang Q, Zou Y (2009) Current status and challenges of rice production in China. Plant Prod Sci 12:3–8
Pradhan A, Naik N, Sahoo KK (2015) RNAi mediated drought and salinity stress tolerance in plants. Am J Plant Sci 6:1990
Rahman KM, Zhang D (2018) Effects of fertilizer broadcasting on the excessive use of inorganic fertilizers and environmental sustainability. Sustainability 10:759
Rahman M, Juraimi AS, Suria J et al (2012) Response of weed flora to different herbicides in aerobic rice system. Sci Res Essays 7:12–23
Rahman MA, Bimpong IK, Bizimana JB et al (2017) Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice 10(1):47. https://doi.org/10.1186/s12284-017-0186-x
Raj SK, Syriac EK (2017) Weed management in direct seeded rice: a review. Agric Rev 38:41–50. https://doi.org/10.18805/ag.v0iOF.7307
Ranawake AL, Manangkil OE, Yoshida S et al (2014) Mapping QTLs for cold tolerance at germination and the early seedling stage in rice (Oryza sativa L.). Biotechnol Biotechnol Equip 28:989–998. https://doi.org/10.1080/13102818.2014.978539
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS One 8:e66428
Ray DK, Gerber JS, MacDonald GK, West PC (2015) Climate variation explains a third of global crop yield variability. Nat Commun 6:1–9
Roy-Barman S, Chattoo BB (2005) Rice blast fungus sequenced. Curr Sci 89:930
Sahebi M, Hanafi MM, Rafii MY et al (2018) Improvement of drought tolerance in rice (Oryza sativa L.): genetics, genomic tools, and the WRKY gene family. Biomed Res Int 2018:3158474
Saika H, Oikawa A, Matsuda F et al (2011) Application of gene targeting to designed mutation breeding of high-tryptophan rice. Plant Physiol 156:1269–1277
Saikumar S, Varma CMK, Saiharini A et al (2016) Grain yield responses to varied level of moisture stress at reproductive stage in an interspecific population derived from Swarna/O. glaberrima introgression line. NJAS Wageningen J Life Sci 78:111–122
Sandhu N, Kumar A (2017) Bridging the rice yield gaps under drought: QTLs, genes, and their use in breeding programs. Agronomy 7:27. https://doi.org/10.3390/agronomy7020027
Schläppi MR, Jackson AK, Eizenga GC et al (2017) Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA mini-core collection. Front Plant Sci 8:957. https://doi.org/10.3389/fpls.2017.00957
Septiningsih EM, Pamplona AM, Sanchez DL et al (2009) Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot 103(2):151–160
Serrano-Mislata A, Bencivenga S, Bush M et al (2017) DELLA genes restrict inflorescence meristem function independently of plant height. Nat Plants 3:749
Shakiba E, Edwards JD, Jodari F et al (2017) Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS One 12:1–22. https://doi.org/10.1371/journal.pone.0172133
Sharma M, Pandey GK (2016) Expansion and function of repeat domain proteins during stress and development in plants. Front Plant Sci 6:1218
Shinada H, Iwata N, Sato T, Fujino K (2014) QTL pyramiding for improving of cold tolerance at fertilization stage in rice. Breed Sci 63:483–488. https://doi.org/10.1270/jsbbs.63.483
Shirasawa K, Takeuchi Y, Ebitani T et al (2008) Identification of gene for rice (Oryza sativa) seed lipoxygenase-3 involved in the generation of stale flavor and development of SNP markers for lipoxygenase-3 deficiency. Breed Sci 58(2):169–176
Shrivastava P, Kumar R (2015) Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci 22:123–131
Singh K, Kumar V, Saharawat YS et al (2013) Weedy rice: an emerging threat for direct-seeded rice production systems in India. J Rice Res 1:1–6. (open access)
Singh H, Jassal RK, Kang JS et al (2015) Seed priming techniques in field crops: a review. Agric Rev 36:251–264
Singh R, Singh Y, Xalaxo S et al (2016) From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci 242:278–287
Sripongpangkul K, Posa GBT, Senadhira DW et al (2000) Genes/QTLs affecting flood tolerance in rice. Theor Appl Genet 101(7):1074–1081
Sui B, Feng X, Tian G et al (2013) Optimizing nitrogen supply increases rice yield and nitrogen use efficiency by regulating yield formation factors. Field Crops Res 150:99–107
Sun J, Yang L, Wang J et al (2018) Identification of a cold-tolerant locus in rice (Oryza sativa L.) using bulked segregant analysis with a next-generation sequencing strategy. Rice 11:24
Suprihatno B, Coffman WR (1981) Inheritance of submergence tolerance in rice (Oryza sativa L.). SABRAO J 13(2):98–108
Swain P, Anumalla M, Prusty S et al (2014) Characterization of some Indian native land race rice accessions for drought tolerance at seedling stage. Aust J Crop Sci 8:324–331
Swamy BPM, Kumar A (2013) Genomics-based precision breeding approaches to improve drought tolerance in rice. Biotechnol Adv 31:1308–1318. https://doi.org/10.1016/j.quaint.2017.02.033
Tigchelaar M, Battisti DS, Naylor RL, Ray DK (2018) Future warming increases probability of globally synchronized maize production shocks. Proc Natl Acad Sci U S A 115:6644–6649
Toojinda T, Siangliw M, Tragoonrung S et al (2003) Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann Bot 91(2):243–253
Tuberosa R (2012) Phenotyping for drought tolerance of crops in the genomics era. Front Physiol 3:347. https://doi.org/10.3389/fphys.2012.00347
United Nations (2019) World population prospects 2019: highlights. United Nations Department of Economic and Social Affairs, New York
Unnevehr LJ, Juliano BO, Perez CM (1985) Consumer demand for rice grain quality in Southeast Asia. International rice research conference, International Rice Research Institute, pp 15–23
Utsumi Y, Utsumi C, Sawada T et al (2011) Functional diversity of isoamylase oligomers: the ISA1 homo-oligomer is essential for amylopectin biosynthesis in rice endosperm. Plant Physiol 156:61–77
Van Oort PAJJ (2018) Mapping abiotic stresses for rice in Africa: drought, cold, iron toxicity, salinity and sodicity. Field Crops Res 219:55–75. https://doi.org/10.1016/j.fcr.2018.01.016
Van Oort PA, Zwart SJ (2018) Impacts of climate change on rice production in Africa and causes of simulated yield changes. Glob Chang Biol 24(3):1029–1045
Vemanna RS, Bakade R, Bharti P et al (2019) Cross-talk signaling in rice during combined drought and bacterial blight stress. Front Plant Sci 10:193
Venu RC, Sreerekha MV, Nobuta K et al (2011) Deep sequencing reveals the complex and coordinated transcriptional regulation of genes related to grain quality in rice cultivars. BMC Genomics 12:190. https://doi.org/10.1111/j.1551-2916.2005.00910.x
Venuprasad R, Dalid CO, Del Valle M et al (2009) Identification and characterization of large-effect quantitative trait loci for grain yield under lowland drought stress in rice using bulk-segregant analysis. Theor Appl Genet 120:177–190
Verma AK, Deepti S (2016) Abiotic stress and crop improvement: current scenario. Adv Plants Agric Res 4:149
Vikram P, Swamy BPM, Dixit S et al (2011) qDTY 1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet 12:89
Vikram P, Swamy BPM, Dixit S et al (2015) Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci Rep 5:1–18. https://doi.org/10.1063/1.4791353
Wang Y, Zhang L, Nafisah A et al (2013) Selection efficiencies for improving drought/salt tolerances and yield using introgression breeding in rice (Oryza sativa L.). Crop J 1:134–142. https://doi.org/10.1016/j.cj.2013.07.006
Wang W, Fu B, Ali J et al (2015) Genome-wide responses to selection and genetic networks underlying submergence tolerance in rice. Plant Genome 8(2):1–13
Wang X, Pang Y, Zhang J et al (2017) Genome-wide and gene-based association mapping for rice eating and cooking characteristics and protein content. Sci Rep 7:1–10. https://doi.org/10.1038/s41598-017-17347-5
Wing RA, Purugganan MD, Zhang Q (2018) The rice genome revolution: from an ancient grain to Green Super Rice. Nat Rev Genet 19:505–517. https://doi.org/10.1038/s41576-018-0024-z
Xie Y, Shen R, Chen L, Liu Y-G (2019) Molecular mechanisms of hybrid sterility in rice. Sci China Life Sci 62(6):737–743
Xing Y, Guo S, Chen X et al (2018) Nitrogen metabolism is affected in the nitrogen-deficient rice mutant esl4 with a calcium-dependent protein kinase gene mutation. Plant Cell Physiol 59:2512–2525
Xu Y, Crouch JH (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48:391–407
Xu K, Xu X, Fukao T et al (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442(7103):705–708
Yadav S, Sandhu N, Singh VK et al (2019) Genotyping-by-sequencing based QTL mapping for rice grain yield under reproductive stage drought stress tolerance. Sci Rep 9:1–12
Yano M, Lin HX, Takeuchi Y et al (2003) Marker-assisted dissection and pyramiding of complex traits in rice. In: Mew TW et al (eds) Rice Science: Innovations and Impact for Livelihood. IRRI, pp 257–263
Ye C, Fukai S, Godwin I et al (2009) Cold tolerance in rice varieties at different growth stages. Crop Pasture Sci 60:328–338
Yorobe J, Pede V, Rejesus R et al (2014) Yield and income effects of the Green Super Rice (GSR) varieties: evidence from a fixed-effects model in the Philippines. Selected paper prepared for presentation at the Agricultural & Applied Economics Association’s 2014 Annual Meeting, Minneapolis, MN, July 27–29, 2014. 31 p. https://doi.org/10.1063/1.1609251
Yorobe JM, Ali J, Pede VO et al (2016) Yield and income effects of rice varieties with tolerance of multiple abiotic stresses: the case of green super rice (GSR) and flooding in the Philippines. Agric Econ 47:1–11
Yu Y, Zhao Z, Shi Y et al (2016) Hybrid sterility in rice (Oryza sativa L.) involves the tetratricopeptide repeat domain containing protein. Genetics 203:1439–1451
Yu S, Ali J, Zhang C et al (2020) Genomic breeding of Green Super Rice varieties and their deployment in Asia and Africa. Theor Appl Genet 133:1427–1442
Yuan LP (2017) Progress in super-hybrid rice breeding. Crop J 5:100W102
Yun BB-W, Kim M-GM, Handoyo T, Kim KK-M (2014) Analysis of rice grain quality-associated quantitative trait loci by using genetic mapping. Am J Plant Sci 5:1125. https://doi.org/10.4236/ajps.2014.59125
Zhang Q (2007) Strategies for developing Green Super Rice. Proc Natl Acad Sci U S A 104:16402–16409. https://doi.org/10.1073/pnas.0708013104
Zhang F, Zhai HQ, Paterson AH et al (2011) Dissecting genetic networks underlying complex phenotypes: the theoretical framework. PLoS One 6(1):e14541
Zhang F, Zhang F, Huang L et al (2016) Overlap between signaling pathways responsive to Xanthomonas oryzae pv. oryzae infection and drought stress in rice introgression line revealed by RNA-seq. J Plant Growth Regul 35:345–356. https://doi.org/10.1007/s00344-015-9538-1
Zhang Z, Li JJJ, Pan Y et al (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat Commun 8:1–13. https://doi.org/10.1007/BF01337500
Zhang J, Guo T, Yang J et al (2020) QTL mapping and haplotype analysis revealed candidate genes for grain thickness in rice (Oryza sativa L.). Mol Breed 40:1–12
Zhou Y, Cai H, Xiao J et al (2009) Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor Appl Genet 118:1381–1390
Zhou Z, Li H, Sun Y et al (2010) Effect of selection for high yield, drought and salinity tolerances on yield-related traits in rice (Oryza sativa L.). Acta Agron Sin 36:1725–1735
Zhu Y, Chen K, Mi X et al (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS One 10:e0145704. https://doi.org/10.1371/journal.pone.0145704
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this chapter
Cite this chapter
Ali, J., Anumalla, M., Murugaiyan, V., Li, Z. (2021). Green Super Rice (GSR) Traits: Breeding and Genetics for Multiple Biotic and Abiotic Stress Tolerance in Rice. In: Ali, J., Wani, S.H. (eds) Rice Improvement. Springer, Cham. https://doi.org/10.1007/978-3-030-66530-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-66530-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66529-6
Online ISBN: 978-3-030-66530-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)