Abstract
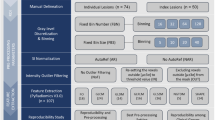
‘Radiomics’ is utilized to improve the prediction of patient overall survival and/or outcome. Target segmentation, feature extraction, feature selection, and classification model are the fundamental blocks of a radiomics workflow. Nevertheless, these blocks can be affected by several issues, i.e. high inter- and intra-observer variability. To overcome these issues obtaining reproducible results, we propose a novel radiomics workflow to identify a relevant prognostic model concerning a real clinical problem. In the specific, we propose an operator-independent segmentation system with the consequent automatic extraction of radiomics features, and a novel feature selection approach to create a relevant predictive model in 46 patients with prostate lesion underwent magnetic resonance imaging.
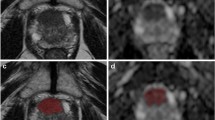
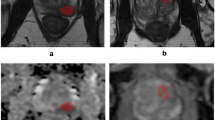
In the specific, using an operator-independent method of target segmentation based on an active contour, ad-hoc automated high-throughput analysis tool capable of calculating a total of 290 radiomics features for each imaging sequence, a novel statistical system for feature reduction and selection, and the discriminant analysis as a method for feature classification, we propose a performant and replicable radiomics workflow for the diagnosis of prostate cancer.
The proposed workflow revealed three and five relevant features on T2-weighted and apparent diffusion coefficient (ADC) maps images, respectively, that were significantly correlated with the histopathological results. In the specific, good performance in lesion discrimination was obtained using the combination of the selected features (accuracy 76.76% and 75.20%, for T2-weighted and ADC maps images, respectively) in an operator-independent and automatic way.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Zhang, Z., Sejdić, E.: Radiological images and machine learning: trends, perspectives, and prospects. Comput. Biol. Med. 108, 354–370 (2019)
Hatt, M., Tixier, F., Visvikis, D., Cheze Le Rest, C.: Radiomics in PET/CT: more than meets the eye? J. Nucl. Med. 58, 365–366 (2017)
Sala, E., Mema, E., Himoto, Y., Veeraraghavan, H., Brenton, J.D., Snyder, A., et al.: Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin. Radiol. 72, 3–10 (2017)
Negrini, S., Gorgoulis, V.G., Halazonetis, T.D.: Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 (2010)
Gerlinger, M., Swanton, C.: How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br. J. Cancer 103, 1139–1143 (2010)
Thawani, R., McLane, M., Beig, N., Ghose, S., Prasanna, P., Velcheti, V., et al.: Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer 2018(115), 134–141 (2017). https://doi.org/10.1016/j.lungcan.2017.10.015
Giambelluca, D., et al.: PI-RADS 3 lesions: role of prostate MRI texture analysis in the identification of prostate cancer. Curr. Probl. Diagn Radiol. (2019, in press). https://doi.org/10.1067/j.cpradiol.2019.10.009
Ugga, L., et al.: Prediction of high proliferative index in pituitary macroadenomas using MRI-based radiomics and machine learning. Neuroradiology 61(12), 1365–1373 (2019)
Stefano, A., et al.: A preliminary PET radiomics study of brain metastases using a fully automatic segmentation method. BMC Bioinform. (2020, in press)
Cuocolo, R., et al.: Prostate MRI technical parameters standardization: a systematic review on adherence to PI-RADSv2 acquisition protocol. Eur. J. Radiol. (2019)
Nioche, C., et al.: Lifex: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 78, 4786–4789 (2018)
Szczypiński, P.M., Strzelecki, M., Materka, A., Klepaczko, A.: MaZda-A software package for image texture analysis. Comput. Methods Programs Biomed. 94, 66–76 (2009)
Fang, Y.H.D., et al.: Development and evaluation of an open-source software package “cGITA” for quantifying tumor heterogeneity with molecular images. Biomed. Res. Int. (2014)
Comelli, A., Agnello, L., Vitabile, S.: An ontology-based retrieval system for mammographic reports. In: Proceedings - IEEE Symposium on Computers and Communications (2016)
Pedregosa, F., et al.: Scikit-learn: machine learning in Python. J. Mach. Learn. Res. (2011)
Cuocolo, R., et al.: Machine learning applications in prostate cancer magnetic resonance imaging. Eur. Radiol. Exp. 3, 35 (2019)
Rizzo, S., et al.: Radiomics: the facts and the challenges of image analysis. Eur. Radiol. Exp. (2018)
Comelli, A., et al.: A kernel support vector machine based technique for Crohn’s disease classification in human patients. In: Barolli, L., Terzo, O. (eds.) CISIS 2017. AISC, vol. 611, pp. 262–273. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-61566-0_25
Litjens, G., et al.: Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med. Image Anal. (2014)
Chandra, S.S., et al.: Patient specific prostate segmentation in 3-D magnetic resonance images. IEEE Trans. Med. Imaging (2012)
Tsai, A., et al.: A shape-based approach to the segmentation of medical imagery using level sets. IEEE Trans. Med. Imaging (2003)
Wang, B., et al.: Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation. Med. Phys. (2019)
Korsager, A.S., et al.: The use of atlas registration and graph cuts for prostate segmentation in magnetic resonance images. Med. Phys. (2015)
Tian, Z., Liu, L., Fei, B.: A fully automatic multi-atlas based segmentation method for prostate MR images. In: Medical Imaging 2015: Image Processing (2015)
Guo, Y., Gao, Y., Shao, Y., Price, T., Oto, A., Shen, D.: Deformable segmentation of 3D MR prostate images via distributed discriminative dictionary and ensemble learning. Med. Phys. (2014)
Toth, R., Madabhushi, A.: Multifeature landmark-free active appearance models: application to prostate MRI segmentation. IEEE Trans. Med. Imaging (2012)
Yang, X., et al.: 3D prostate segmentation in MR image using 3D deeply supervised convolutional neural networks. Med. Phys. (2018)
Jia, H., Xia, Y., Song, Y., Cai, W., Fulham, M., Feng, D.D.: Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing (2018)
Comelli, A., et al.: A smart and operator independent system to delineate tumours in positron emission tomography scans. Comput. Biol. Med. 102, 1–15 (2018). https://doi.org/10.1016/j.compbiomed.2018.09.002
Comelli, A., Stefano, A.: A fully automated segmentation system of positron emission tomography studies. In: Zheng, Y., Williams, B.M., Chen, K. (eds.) MIUA 2019. CCIS, vol. 1065, pp. 353–363. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-39343-4_30
Lankton, S., Nain, D., Yezzi, A., Tannenbaum, A.: Hybrid geodesic region-based curve evolutions for image segmentation. In: Hsieh, J., Flynn, M.J. (eds.) Medical Imaging 2007: Physics of Medical Imaging. International Society for Optics and Photonics, p. 65104U (2007). https://doi.org/10.1117/12.709700
Cohen, J., Cohen, P., West, S., Aiken, L.: Applied Multiple Regression/Correlation Analysis for the Behavioral Sciences, 2nd edn. Lawrence Erlbaum Associates, Hillsdale (1983)
Comelli, A., et al.: Active contour algorithm with discriminant analysis for delineating tumors in positron emission tomography. Artif. Intell. Med. 94, 67–78 (2019). https://doi.org/10.1016/j.artmed.2019.01.002
Comelli, A., Stefano, A., Benfante, V., Russo, G.: Normal and abnormal tissue classification in positron emission tomography oncological studies. Pattern Recogn. Image Anal. 28, 106–113 (2018). https://doi.org/10.1134/S1054661818010054
Armand, S., Watelain, E., Roux, E., Mercier, M., Lepoutre, F.X.: Linking clinical measurements and kinematic gait patterns of toe-walking using fuzzy decision trees. Gait Posture. 25, 475–484 (2007)
Comelli, A., et al.: Development of a new fully three-dimensional methodology for tumours delineation in functional images. Comput. Biol. Med. 120, 103701 (2020). https://doi.org/10.1016/j.compbiomed.2020.103701
Comelli, A., et al.: K-nearest neighbor driving active contours to delineate biological tumor volumes. Eng. Appl. Artif. Intell. 81, 133–144 (2019). https://doi.org/10.1016/j.engappai.2019.02.005
Comelli, A., et al.: Tissue Classification to Support Local Active Delineation of Brain Tumors. In: Zheng, Y., Williams, B.M., Chen, K. (eds.) MIUA 2019. CCIS, vol. 1065, pp. 3–14. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-39343-4_1
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Comelli, A. et al. (2020). Radiomics: A New Biomedical Workflow to Create a Predictive Model. In: Papież, B., Namburete, A., Yaqub, M., Noble, J. (eds) Medical Image Understanding and Analysis. MIUA 2020. Communications in Computer and Information Science, vol 1248. Springer, Cham. https://doi.org/10.1007/978-3-030-52791-4_22
Download citation
DOI: https://doi.org/10.1007/978-3-030-52791-4_22
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-52790-7
Online ISBN: 978-3-030-52791-4
eBook Packages: Computer ScienceComputer Science (R0)