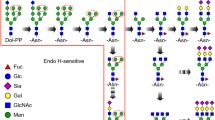
Abstract
Glycoproteins result from post-translational modification of proteins by glycans attached to certain side chains, with possible heterogeneity due to different structures being possible at the same glycosylation site.
In contrast to the mammalian systems, analysis of invertebrate glycans presents a challenge in analysis as there exist unfamiliar epitopes and a high degree of structural and isomeric variation between different species—Caenorhabditis elegans is no exception. Simple screening using lectins and antibodies can yield hints regarding which glycan epitopes are present in wild-type and mutant strains, but detailed analysis is necessary for determining more exact glycomic information. Here, our analytical approach is to analyze N- and O-glycans involving “off-line” RP-HPLC MALDI-TOF MS/MS. Enrichment and labeling steps facilitate the analysis of single structures and provide isomeric separation. Thereby, the “simple” worm expresses over 200 N-glycan structures varying depending on culture conditions or the genetic background.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- DTT:
-
dithiothreitol
- HRP:
-
horseradish peroxidase
- MALDI-TOF MS:
-
matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry
- NPGC:
-
non-porous graphitized carbon
- PA:
-
pyridylamino
- PC:
-
phosphorylcholine
- RP-HPLC:
-
reversed phase high-pressure liquid chromatography
- SDS-PAGE:
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
References
Varki A (2011) Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells. Cold Spring Harb Perspect Biol 3(6):a005462. https://doi.org/10.1101/cshperspect.a005462
Spiro RG (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R–56R
Aebi M (2013) N-linked protein glycosylation in the ER. Biochim Biophys Acta 1833(11):2430–2437. https://doi.org/10.1016/j.bbamcr.2013.04.001
Schiller B, Hykollari A, Yan S et al (2012) Complicated N-linked glycans in simple organisms. Biol Chem Hoppe Seyler 393:661–673
Eckmair B, Jin C, Abed-Navandi D et al (2016) Multi-step fractionation and mass spectrometry reveals zwitterionic and anionic modifications of the N- and O-glycans of a marine snail. Mol Cell Proteomics 15:573–597. https://doi.org/10.1074/mcp.M115.051573
Stanton R, Hykollari A, Eckmair B et al (2017) The underestimated N-glycomes of lepidopteran species. Biochim Biophys Acta 1861(4):699–714. https://doi.org/10.1016/j.bbagen.2017.01.009
Yan S, Vanbeselaere J, Jin C et al (2018) Core richness of N-glycans of Caenorhabditis elegans: a case study on chemical and enzymatic release. Anal Chem 90(1):928–935. https://doi.org/10.1021/acs.analchem.7b03898
Hirabayashi J, Hayama K, Kaji H et al (2002) Affinity capturing and gene assignment of soluble glycoproteins produced by the nematode Caenorhabditis elegans. J Biochem (Tokyo) 132(1):103–114. https://doi.org/10.1093/oxfordjournals.jbchem.a003186
Fan X, She YM, Bagshaw RD et al (2005) Identification of the hydrophobic glycoproteins of Caenorhabditis elegans. Glycobiology 15(10):952–964. https://doi.org/10.1093/glycob/cwi075
Shimizu T, Kato Y, Sakai Y et al (2019) N-glycosylation of the Discoidin domain receptor is required for axon regeneration in Caenorhabditis elegans. Genetics 213(2):491–500. https://doi.org/10.1534/genetics.119.302492
Rahman M, Ramirez-Suarez NJ, Diaz-Balzac CA et al (2022) Specific N-glycans regulate an extracellular adhesion complex during somatosensory dendrite patterning. EMBO Rep 23(7):e54163. https://doi.org/10.15252/embr.202154163
Roberts B, Antonopoulos A, Haslam SM et al (2013) Novel expression of Haemonchus contortus vaccine candidate aminopeptidase H11 using the free-living nematode Caenorhabditis elegans. Vet Res 44:111. https://doi.org/10.1186/1297-9716-44-111
Hykollari A, Malzl D, Yan S et al (2017) Hydrophilic interaction anion exchange for separation of multiply modified neutral and anionic Dictyostelium N-glycans. Electrophoresis 38:2175–2183. https://doi.org/10.1002/elps.201700073
Paschinger K, Wilson IBH (2016) Analysis of zwitterionic and anionic N-linked glycans from invertebrates and protists by mass spectrometry. Glycoconj J 33:273–283. https://doi.org/10.1007/s10719-016-9650-x
Paschinger K, Wöls F, Yan S et al (2023) N-glycan antennal modifications are altered in Caenorhabditis elegans lacking the HEX-4 N-acetylgalactosamine-specific hexosaminidase. J Biol Chem 299:103053. https://doi.org/10.1016/j.jbc.2023.103053
Wilson IBH, Yan S, Jin C et al (2023) Increasing complexity of the N-glycome during Caenorhabditis development. Mol Cell Proteomics 22:100505. https://doi.org/10.1016/j.mcpro.2023.100505
Vanbeselaere J, Yan S, Joachim A et al (2018) The parasitic nematode Oesophagostomum dentatum synthesizes unusual glycosaminoglycan-like O-glycans. Glycobiology 28(7):474–481. https://doi.org/10.1093/glycob/cwy045
Paschinger K, Gonzalez-Sapienza GG, Wilson IBH (2012) Mass spectrometric analysis of the immunodominant glycan epitope of Echinococcus granulosus antigen Ag5. Int J Parasitol 42(3):279–285. https://doi.org/10.1016/j.ijpara.2012.01.002
Hykollari A, Malzl D, Eckmair B et al (2018) Isomeric separation and recognition of anionic and zwitterionic N-glycans from royal jelly glycoproteins. Mol Cell Proteomics 17(11):2177–2196. https://doi.org/10.1074/mcp.RA117.000462
Hykollari A, Malzl D, Wilson IBH et al (2019) Protein-specific analysis of invertebrate glycoproteins. Methods Mol Biol 1871:421–435. https://doi.org/10.1007/978-1-4939-8814-3_24
Iskratsch T, Braun A, Paschinger K et al (2009) Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal Biochem 386(2):133–146. https://doi.org/10.1016/j.ab.2008.12.005
Mikolajek H, Kolstoe SE, Pye VE et al (2011) Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component. J Mol Recognit 24(2):371–377. https://doi.org/10.1002/jmr.1090
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77(1):71–94
Lewis JA, Fleming JT (1995) Basic culture methods. Methods Cell Biol 48:3–29
Hykollari A, Paschinger K, Eckmair B et al (2017) Analysis of invertebrate and Protist N-Glycans. Methods Mol Biol 1503:167–184. https://doi.org/10.1007/978-1-4939-6493-2_13
Dragosits M, Pflugl S, Kurz S et al (2014) Recombinant aspergillus β-galactosidases as a robust glycomic and biotechnological tool. Appl Microbiol Biotechnol 98(8):3553–3567. https://doi.org/10.1007/s00253-013-5192-3
Dragosits M, Yan S, Razzazi-Fazeli E et al (2015) Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology 25(4):448–464. https://doi.org/10.1093/glycob/cwu132
Kameyama A, Thet Tin WW, Toyoda M et al (2019) A practical method of liberating O-linked glycans from glycoproteins using hydroxylamine and an organic superbase. Biochem Biophys Res Commun 513(1):186–192. https://doi.org/10.1016/j.bbrc.2019.03.144
Jiménez-Castells C, Vanbeselaere J, Kohlhuber S et al (2017) Gender and developmental specific N-glycomes of the porcine parasite Oesophagostomum dentatum. Biochim Biophys Acta 1861(2):418–430. https://doi.org/10.1016/j.bbagen.2016.10.011
Purohit S, Li T, Guan W et al (2018) Multiplex glycan bead array for high throughput and high content analyses of glycan binding proteins. Nat Commun 9(1):258. https://doi.org/10.1038/s41467-017-02747-y
Paschinger K, Rendić D, Wilson IBH (2009) Revealing the anti-HRP epitope in Drosophila and Caenorhabditis. Glycoconj J 26:385–395
Paschinger K, Gonzalez-Sapienza GG, Wilson IBH (2012) Mass spectrometric analysis of the immunodominant glycan epitope of Echinococcus granulosus antigen Ag5. Int J Parasitol 42:279–285
Aoki K, Perlman M, Lim JM et al (2007) Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem 282:9127–9142. https://doi.org/10.1074/jbc.M606711200
Miyata S, Sato C, Kumita H et al (2006) Flagellasialin: a novel sulfated α2,9-linked polysialic acid glycoprotein of sea urchin sperm flagella. Glycobiology 16(12):1229–1241. https://doi.org/10.1093/glycob/cwl036
Martini F, Eckmair B, Neupert C et al (2019) Highly modified and immunoactive N-glycans of the canine heartworm. Nat Commun 10:75. https://doi.org/10.1038/s41467-018-07948-7
Tsai PL, Chen SF (2017) A brief review of bioinformatics tools for glycosylation analysis by mass spectrometry. Mass Spectrom 6:S0064. https://doi.org/10.5702/massspectrometry.S0064
York WS, Agravat S, Aoki-Kinoshita KF et al (2014) MIRAGE: the minimum information required for a glycomics experiment. Glycobiology 24(5):402–406. https://doi.org/10.1093/glycob/cwu018
Varki A, Cummings RD, Aebi M et al (2015) Symbol nomenclature for graphical representations of Glycans. Glycobiology 25(12):1323–1324. https://doi.org/10.1093/glycob/cwv091
Morelle W, Faid V, Chirat F et al (2009) Analysis of N- and O-linked glycans from glycoproteins using MALDI-TOF mass spectrometry. Methods Mol Biol 534:5–21. https://doi.org/10.1007/978-1-59745-022-5_1
Mereiter S, Magalhaes A, Adamczyk B et al (2016) Glycomic and sialoproteomic data of gastric carcinoma cells overexpressing ST3GAL4. Data Brief 7:814–833. https://doi.org/10.1016/j.dib.2016.03.022
Acknowledgments
This work was supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF; grants P32572 and P29466 to K.P and I.B.H.W.).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Paschinger, K., Vanbeselaere, J., Wilson, I.B.H. (2024). Analysis of Caenorhabditis Protein Glycosylation. In: Bradfute, S.B. (eds) Recombinant Glycoproteins. Methods in Molecular Biology, vol 2762. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3666-4_8
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3666-4_8
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3665-7
Online ISBN: 978-1-0716-3666-4
eBook Packages: Springer Protocols