Abstract
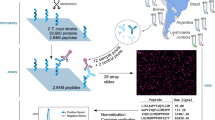
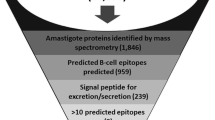
Chagas disease, caused by the protozoan parasite Trypanosoma cruzi, is considered a Neglected Tropical Disease. Limited investment is assigned to its study and control, even though it is one of the most prevalent parasitic infections worldwide. An innovative vaccination strategy involving an epitope-based vaccine that displays multiple immune determinants originating from different antigens could counteract the high biological complexity of the parasite and lead to a wide and protective immune response. In this chapter, we describe a computational reverse vaccinology pipeline applied to identify the most promising peptide sequences from T. cruzi proteins, prioritizing evolutionary conserved sequences, to finally select a list of T and B cell epitope candidates to be further tested in an experimental setting.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Beaumier CM, Gillespie PM, Strych U et al (2016) Status of vaccine research and development of vaccines for Chagas disease. Vaccine 34:2996–3000. https://doi.org/10.1016/j.vaccine.2016.03.074
Pinazo M-J, Gascon J (2015) Chagas disease: from Latin America to the world. Rep Parasitol 4:7–14. https://doi.org/10.2147/RIP.S57144
Gascon J, Bern C, Pinazo M-J (2010) Chagas disease in Spain, the United States and other non-endemic countries. Acta Trop 115:22–27. https://doi.org/10.1016/j.actatropica.2009.07.019
Aldasoro E, Posada E, Requena-Méndez A et al (2018) What to expect and when: benznidazole toxicity in chronic Chagas’ disease treatment. J Antimicrob Chemother 73:1060–1067. https://doi.org/10.1093/jac/dkx516
Jackson Y, Alirol E, Getaz L et al (2010) Tolerance and safety of nifurtimox in patients with chronic Chagas disease. Clin Infect Dis 51:e69–e75. https://doi.org/10.1086/656917
Teh-Poot C, Dumonteil E (2019) Mining Trypanosoma cruzi genome sequences for antigen discovery and vaccine development. In: Gómez KA, Buscaglia CA (eds) T. cruzi infection. Springer New York, New York, pp 23–34
Vita R, Mahajan S, Overton JA et al (2019) The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47:D339–D343. https://doi.org/10.1093/nar/gky1006
Galanis KA, Nastou KC, Papandreou NC et al (2021) Linear B-cell epitope prediction for in silico vaccine design: a performance review of methods available via command-line interface. Int J Mol Sci 22:3210. https://doi.org/10.3390/ijms22063210
Sullivan NL, Eickhoff CS, Sagartz J, Hoft DF (2015) Deficiency of antigen-specific B cells results in decreased Trypanosoma cruzi systemic but not mucosal immunity due to CD8 T cell exhaustion. J Immunol 194:1806–1818. https://doi.org/10.4049/jimmunol.1303163
Buschiazzo A, Muiá R, Larrieux N et al (2012) Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors. PLoS Pathog 8:e1002474. https://doi.org/10.1371/journal.ppat.1002474
Varadi M, Anyango S, Deshpande M et al (2022) AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50:D439–D444. https://doi.org/10.1093/nar/gkab1061
Aslett M, Aurrecoechea C, Berriman M et al (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38:D457–D462. https://doi.org/10.1093/nar/gkp851
Amos B, Aurrecoechea C, Barba M et al (2022) VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res 50:D898–D911. https://doi.org/10.1093/nar/gkab929
The NIH HMP Working Group, Peterson J, Garges S et al (2009) The NIH human microbiome project. Genome Res 19:2317–2323. https://doi.org/10.1101/gr.096651.109
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Nielsen M, Lundegaard C, Lund O, Keşmir C (2005) The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33–41. https://doi.org/10.1007/s00251-005-0781-7
Besser H, Louzoun Y (2018) Cross-modality deep learning-based prediction of TAP binding and naturally processed peptide. Immunogenetics 70:419–428. https://doi.org/10.1007/s00251-018-1054-6
Reynisson B, Alvarez B, Paul S et al (2020) NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48:W449–W454. https://doi.org/10.1093/nar/gkaa379
Teufel F, Almagro Armenteros JJ, Johansen AR et al (2022) SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol 40:1023–1025. https://doi.org/10.1038/s41587-021-01156-3
Gíslason MH, Nielsen H, Almagro Armenteros JJ, Johansen AR (2021) Prediction of GPI-anchored proteins with pointer neural networks. Curr Res Biotechnol 3:6–13. https://doi.org/10.1016/j.crbiot.2021.01.001
Jespersen MC, Peters B, Nielsen M, Marcatili P (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45:W24–W29. https://doi.org/10.1093/nar/gkx346
Hubbard SJ, Thornton JM (1993) ‘NACCESS’, computer program. Department of Biochemistry and Molecular Biology, University College, London
Altschul S (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Resource Coordinators NCBI, Agarwala R, Barrett T et al (2018) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13. https://doi.org/10.1093/nar/gkx1095
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:623–656. https://doi.org/10.1002/j.1538-7305.1948.tb00917.x
Alonso-Padilla J, Lafuente EM, Reche PA (2017) Computer-aided design of an epitope-based vaccine against Epstein-Barr virus. J Immunol Res 2017:1–15. https://doi.org/10.1155/2017/9363750
Stewart JJ, Lee CY, Ibrahim S et al (1997) A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol Immunol 34:1067–1082. https://doi.org/10.1016/S0161-5890(97)00130-2
Weiskopf D, Angelo MA, de Azeredo EL et al (2013) Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci U S A 110:E2046–E2053. https://doi.org/10.1073/pnas.1305227110
Greenbaum J, Sidney J, Chung J et al (2011) Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 63:325–335. https://doi.org/10.1007/s00251-011-0513-0
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Tunyasuvunakool K, Adler J, Wu Z et al (2021) Highly accurate protein structure prediction for the human proteome. Nature 596:590–596. https://doi.org/10.1038/s41586-021-03828-1
Petersen B, Petersen TN, Andersen P et al (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 9:51. https://doi.org/10.1186/1472-6807-9-51
Acknowledgments
We acknowledge support from the Spanish Ministry of Science and Innovation and State Research Agency through the “Centro de Excelencia Severo Ochoa 2019-2023” Program (CEX2018-000806-S), and support from the Generalitat de Catalunya through the CERCA Program. This research was also supported by CIBER—Consorcio Centro de Investigación Biomédica en Red- (CB 2021), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea—NextGenerationEU. A.R-L., J.G. and J.A-P. belong to the AGAUR Grup de Recerca Consolidat 2021SGR01562.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Ros-Lucas, A., Rioja-Soto, D., Gascón, J., Alonso-Padilla, J. (2023). Computational Prediction of Trypanosoma cruzi Epitopes Toward the Generation of an Epitope-Based Vaccine Against Chagas Disease. In: Reche, P.A. (eds) Computational Vaccine Design. Methods in Molecular Biology, vol 2673. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3239-0_32
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3239-0_32
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3238-3
Online ISBN: 978-1-0716-3239-0
eBook Packages: Springer Protocols