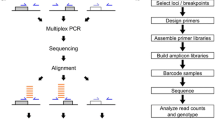
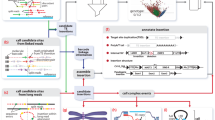
Abstract
Transposable element (TE) insertions are a major source of structural variation in the human genome. Due to the repetitive nature and biological importance of TEs, many bioinformatic tools have been developed to identify and genotype TE insertion polymorphisms using high-throughput short-reads. In this chapter, we outline recently developed methods to characterize TE insertion polymorphisms in human populations. We also provide detailed protocols to tackle this question primarily using three software: MELT2, ERVcaller, and TypeREF.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bourque G, Burns KH, Gehring M et al (2018) Ten things you should know about transposable elements. Genome Biol 19:199
Zook JM, Hansen NF, Olson ND et al (2020) A robust benchmark for detection of germline large deletions and insertions. Nat Biotechnol 38:1347–1355
Groza C, Chen X, Pacis A et al (2021) Genome graphs detect human polymorphisms in active epigenomic states during influenza infection. https://doi.org/10.1101/2021.09.29.462206
Goubert C, Zevallos NA, Feschotte C (2020) Contribution of unfixed transposable element insertions to human regulatory variation. Philos Trans R Soc Lond Ser B Biol Sci 375:20190331
Kazazian HH Jr, Moran JV (2017) Mobile DNA in health and disease. N Engl J Med 377:361–370
Payer LM, Burns KH (2019) Transposable elements in human genetic disease. Nat Rev Genet 20:760–772
Tang Z, Steranka JP, Ma S et al (2017) Human transposon insertion profiling: analysis, visualization and identification of somatic LINE-1 insertions in ovarian cancer. Proc Natl Acad Sci U S A 114:E733–E740
Karamitros T, Hurst T, Marchi E et al (2018) Human Endogenous Retrovirus-K HML-2 integration within RASGRF2 is associated with intravenous drug abuse and modulates transcription in a cell-line model. Proc Natl Acad Sci U S A 115:10434–10439
Wildschutte JH, Williams ZH, Montesion M et al (2016) Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci U S A 113:E2326–E2334
Watkins WS, Feusier JE, Thomas J et al (2020) The Simons Genome Diversity Project: a global analysis of mobile element diversity. Genome Biol Evol 12:779–794
Chu C, Borges-Monroy R, Viswanadham VV et al (2021) Comprehensive identification of transposable element insertions using multiple sequencing technologies. Nat Commun 12:3836
Gardner EJ, Lam VK, Harris DN et al (2017) The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res 27:1916–1929
Sudmant PH, Rausch T, Gardner EJ et al (2015) An integrated map of structural variation in 2,504 human genomes. Nature 526:75–81
Stewart C, Kural D, Strömberg MP et al (2011) A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet 7:e1002236
Rishishwar L, Mariño-Ramírez L, Jordan IK (2016) Benchmarking computational tools for polymorphic transposable element detection. Brief Bioinform bbw072
Vendrell-Mir P, Barteri F, Merenciano M et al (2019) A benchmark of transposon insertion detection tools using real data. Mob DNA 10:53
Byrska-Bishop M, Evani US, Zhao X, et al (2021) High coverage whole genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios, https://doi.org/10.1101/2021.02.06.430068
Niu Y, Teng X, Zhou H et al (2022) Characterizing mobile element insertions in 5675 genomes. Nucleic Acids Res 50:2493–2508
Kojima S, Koyama S, Ka M et al (2022) Mobile elements in human population-specific genome and phenotype divergence. https://doi.org/10.1101/2022.03.25.485726
Xue B, Sechi LA, Kelvin DJ (2020) Human endogenous retrovirus K (HML-2) in health and disease. Front Microbiol 11:1690
Feusier J, Watkins WS, Thomas J et al (2019) Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res 29:1567–1577
Ostertag EM, Kazazian HH Jr (2001) Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res 11:2059–2065
Meyer TJ, Srikanta D, Conlin EM et al (2010) Heads or tails: L1 insertion-associated 5′ homopolymeric sequences. Mob DNA 1:7
Mager DL, Goodchild NL (1989) Homologous recombination between the LTRs of a human retrovirus-like element causes a 5-kb deletion in two siblings. Am J Hum Genet 45:848–854
Thomas J, Perron H, Feschotte C (2018) Variation in proviral content among human genomes mediated by LTR recombination. Mob DNA 9:36
Yu T, Huang X, Dou S et al (2021) A benchmark and an algorithm for detecting germline transposon insertions and measuring de novo transposon insertion frequencies. Nucleic Acids Res 49:e44
Bowles H, Kabiljo R, Jones A et al (2022) An assessment of bioinformatics tools for the detection of human endogenous retroviral insertions in short-read genome sequencing data. https://doi.org/10.1101/2022.02.18.481042
Chen X, Li D (2019) ERVcaller: identifying polymorphic endogenous retrovirus and other transposable element insertions using whole-genome sequencing data. Bioinformatics 35:3913–3922
Goubert C, Thomas J, Payer LM et al (2020) TypeTE: a tool to genotype mobile element insertions from whole genome resequencing data. Nucleic Acids Res 48:e36
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993
Di Tommaso P, Chatzou M, Floden EW et al (2017) Nextflow enables reproducible computational workflows. Nat Biotechnol 35:316–319
Mitra-Behura S, Fiolka RP, Daetwyler S (2022) Singularity containers improve reproducibility and ease of use in computational image analysis workflows. Front Bioinform 1
Merkel D (2014) Docker: lightweight linux containers for consistent development and deployment. 2
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, http://arxiv.org/abs/1303.3997
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Zook JM, Hansen NF, Olson ND et al (2019) A robust benchmark for germline structural variant detection. https://doi.org/10.1101/664623
Boissinot S, Davis J, Entezam A et al (2006) Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A 103:9590–9594
Cordaux R, Lee J, Dinoso L et al (2006) Recently integrated Alu retrotransposons are essentially neutral residents of the human genome. Gene 373:138–144
Rishishwar L, Tellez Villa CE, Jordan IK (2015) Transposable element polymorphisms recapitulate human evolution. Mob DNA 6:21
Doronina L, Reising O, Clawson H et al (2019) True homoplasy of retrotransposon insertions in primates. Syst Biol 68:482–493
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Puurand T, Kukuškina V, Pajuste F-D et al (2019) AluMine: alignment-free method for the discovery of polymorphic Alu element insertions. Mob DNA 10:31
Rajaby R, Sung W-K (2018) TranSurVeyor: an improved database-free algorithm for finding non-reference transpositions in high-throughput sequencing data. Nucleic Acids Res 46:e122
Santander CG, Gambron P, Marchi E et al (2017) STEAK: a specific tool for transposable elements and retrovirus detection in high-throughput sequencing data. Virus Evol 3:vex023
Bogaerts-Márquez M, Barrón MG, Fiston-Lavier A-S et al (2019) T-lex3: an accurate tool to genotype and estimate population frequencies of transposable elements using the latest short-read whole genome sequencing data. Bioinformatics 36(4):1191
Zhuang J, Wang J, Theurkauf W et al (2014) TEMP: a computational method for analyzing transposable element polymorphism in populations. Nucleic Acids Res 42:6826–6838
Wildschutte JH, Baron A, Diroff NM et al (2015) Discovery and characterization of Alu repeat sequences via precise local read assembly. Nucleic Acids Res 43:10292–10307
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Chen, X., Bourque, G., Goubert, C. (2023). Genotyping of Transposable Element Insertions Segregating in Human Populations Using Short-Read Realignments. In: Branco, M.R., de Mendoza Soler, A. (eds) Transposable Elements. Methods in Molecular Biology, vol 2607. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2883-6_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2883-6_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2882-9
Online ISBN: 978-1-0716-2883-6
eBook Packages: Springer Protocols