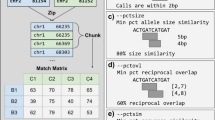
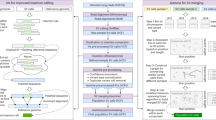
Abstract
We present snakeSV, an open-source fast and scalable framework to analyze genomic structural variation (SV) at scale. The framework is easily deployable using Bioconda and can leverage cluster environments to speed up data processing via parallelization. Providing a set of preconfigured tools, all available at the Bioconda channel for easy installation, snakeSV combines a set of auxiliary scripts that makes it easy to integrate novel tools and features. Execution starts with one or many BAM files and produces a VCF file with SVs detected and jointly genotyped across samples and a report with relevant annotations. We also present two use cases to illustrate the pipeline features to improve SV discovery by using a panel of high-quality SVs and incorporating custom annotations to help biological interpretation.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Collins RL, Brand H, Karczewski KJ et al (2020) A structural variation reference for medical and population genetics. Nature 581:444–451
Sudmant PH, Rausch T, Gardner EJ et al (2015) An integrated map of structural variation in 2,504 human genomes. Nature 526:75–81
Handsaker RE, Van Doren V, Berman JR et al (2015) Large multiallelic copy number variations in humans. Nat Genet 47:296–303
Vialle RA, de Paiva Lopes K, Bennett DA et al (2021) The impact of genomic structural variation on the transcriptome, chromatin, and proteome in the human brain. medRxiv:2021.02.25.21252245
Chiang C, Scott AJ, Davis JR et al (2017) The impact of structural variation on human gene expression. Nat Genet 49:692–699
Han L, Zhao X, Benton ML et al (2020) Functional annotation of rare structural variation in the human brain. Nat Commun 11:2990
Jakubosky D, D’Antonio M, Bonder MJ et al (2020) Properties of structural variants and short tandem repeats associated with gene expression and complex traits. Nat Commun 11:2927
Lupiáñez DG, Kraft K, Heinrich V et al (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025
Collins RL, Brand H, Redin CE et al (2017) Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18:36
Cook EH Jr, Scherer SW (2008) Copy-number variations associated with neuropsychiatric conditions. Nature 455:919–923
Zarrei M, Burton CL, Engchuan W et al (2019) A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom Med 4:26
McCarthy SE, Makarov V, Kirov G et al (2009) Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 41:1223–1227
Sekar A, Bialas AR, de Rivera H et al (2016) Schizophrenia risk from complex variation of complement component 4. Nature 530:177–183
Marshall CR, Howrigan DP, Merico D et al (2017) Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49:27–35
Pinto D, Pagnamenta AT, Klei L et al (2010) Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466:368–372
Sebat J, Lakshmi B, Malhotra D et al (2007) Strong association of de novo copy number mutations with autism. Science 316:445–449
Mitra I, Huang B, Mousavi N et al (2021) Patterns of de novo tandem repeat mutations and their role in autism. Nature 589:246–250
Männik K, Mägi R, Macé A et al (2015) Copy number variations and cognitive phenotypes in unselected populations. JAMA 313:2044–2054
Stefansson H, Meyer-Lindenberg A, Steinberg S et al (2014) CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505:361–366
Abel HJ, Larson DE, Regier AA et al (2020) Mapping and characterization of structural variation in 17,795 human genomes. Nature 583:83–89
Ebert P, Audano PA, Zhu Q et al (2021) Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372:eabf7117
Abyzov A, Urban AE, Snyder M et al (2011) CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21:974–984
Rausch T, Zichner T, Schlattl A et al (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28:i333–i339
Layer RM, Chiang C, Quinlan AR et al (2014) LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15:1–19
Mohiyuddin M, Mu JC, Li J et al (2015) MetaSV: an accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics 31:2741–2744
Becker T, Lee W-P, Leone J et al (2018) FusorSV: an algorithm for optimally combining data from multiple structural variation detection methods. Genome Biol 19:38
Köster J, Rahmann S (2012) Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28:2520–2522
Zook JM, Hansen NF, Olson ND et al (2020) A robust benchmark for detection of germline large deletions and insertions. Nat Biotechnol 38:1347–1355
Nott A, Holtman IR, Coufal NG et al (2019) Brain cell type–specific enhancer–promoter interactome maps and disease-risk association. Science 366:1134–1139
Chen X, Schulz-Trieglaff O, Shaw R et al (2016) Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32:1220–1222
Pedersen B, Layer R, Quinlan AR (2020) smoove: structural-variant calling and genotyping with existing tools. In: Github. https://github.com/brentp/smoove. Accessed 01 Mar 2022
Jeffares DC, Jolly C, Hoti M et al (2017) Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8:14061
Heller D, Vingron M (2019) SVIM: structural variant identification using mapped long reads. Bioinformatics 35:2907–2915
Eggertsson HP, Kristmundsdottir S, Beyter D et al (2019) GraphTyper2 enables population-scale genotyping of structural variation using pangenome graphs. Nat Commun 10:5402
Stone M, Collins R (2016) svtk: Structural variation toolkit. In: Github. https://github.com/talkowski-lab/svtk. Accessed 01 Mar 2022
Heller D, Vingron M (2020) SVIM-asm: structural variant detection from haploid and diploid genome assemblies. Bioinformatics
English AC, Menon VK, Gibbs R, Metcalf GA, Sedlazeck FJ (2022) Truvari: Refined structural variant comparison preserves allelic diversity. bioRxiv 2022.02.21.481353
Gardner EJ, Lam VK, Harris DN et al (2017) The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res 27:1916–1929
The 1000 Genomes Project Consortium (2015) A global reference for human genetic variation. Nature 526:68–74
Kuzniar A, Maassen J, Verhoeven S et al (2020) sv-callers: a highly portable parallel workflow for structural variant detection in whole-genome sequence data. PeerJ 8:e8214
Zarate S, Carroll A, Mahmoud M et al (2020) Parliament2: accurate structural variant calling at scale. Gigascience 9:giaa145
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Vialle, R.A., Raj, T. (2022). snakeSV: Flexible Framework for Large-Scale SV Discovery. In: Proukakis, C. (eds) Genomic Structural Variants in Nervous System Disorders. Neuromethods, vol 182. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2357-2_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2357-2_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2356-5
Online ISBN: 978-1-0716-2357-2
eBook Packages: Springer Protocols