Abstract
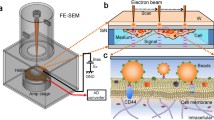
Scanning electron-assisted dielectric microscopy (SE-ADM) is a new microscope technology developed to observe the fine structure of biological samples in aqueous solution. One main advantage of SE-ADM is that it does not require sample pretreatment, including dehydration, drying, and staining, which is indispensable in conventional scanning electron microscopy (SEM) and can cause sample deformation. In addition, the sample is not directly irradiated with an electron beam in SE-ADM, further avoiding damage. The resolution of SE-ADM is higher than that of an optical microscope, which is typically used for observing biological samples in a solution, allowing for the observation of the detailed structure of samples. Considering these advantages, we applied SE-ADM to observe aggregates of therapeutic immunoglobulin G (IgG) of various sizes and shapes in an aqueous solution. In this chapter, we outline the step-by-step procedure for observing aggregates of monoclonal antibodies using SE-ADM and the subsequent analysis of the particle distribution and calculation of the fractal dimension using SE-ADM image data. The proposed method for particle analysis is highly reliable with respect to size measurement and can determine the diameter of a sample with an accuracy of ±20%, a precision of ±10%, and a lower limit of quantification of ≤50 nm. Further, by calculating the fractal dimension of the image, it is possible to classify the shape of the aggregates and determine the mechanism of aggregation.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Guidance for Industry. Immunogenicity Assessment for Therapeutic Protein Products. (2014) Food and Drug Administration
Ecker DM, Jones SD, Levine HL (2015) The therapeutic monoclonal antibody market. mAbs 7:9–14
Elgundi Z, Reslan M, Cruz E et al (2017) The state-of-play and future of antibody therapeutics. Adv Drug Deliv Rev 122:2–19
Wang W, Nema S, Teagarden D (2010) Protein aggregation--pathways and influencing factors. Int J Pharm 390:89–99
Joubert MK, Luo Q, Nashed-Samuel Y et al (2011) Classification and characterization of therapeutic antibody aggregates. J Biol Chem 286:25118–25133
Luo Q, Joubert MK, Stevenson R et al (2011) Chemical modifications in therapeutic protein aggregates generated under different stress conditions. J Biol Chem 286:25134–25144
Correia IR (2010) Stability of IgG isotypes in serum. mAbs 2:221–232
Volkin DB, Hershenson S, Ho RJ et al (2015) Two decades of publishing excellence in pharmaceutical biotechnology. J Pharm Sci 104:290–300
Moussa EM, Panchal JP, Moorthy BS et al (2016) Immunogenicity of therapeutic protein aggregates. J Pharm Sci 105:417–430
Filipe V, Jiskoot W, Basmeleh AH et al (2012) Immunogenicity of different stressed IgG monoclonal antibody formulations in immune tolerant transgenic mice. mAbs 4:740–752
Imamura H, Honda S (2016) Kinetics of antibody aggregation at neutral pH and ambient temperatures triggered by temporal exposure to acid. J Phys Chem B 120:9581–9589
Imamura H, Sasaki A, Honda S (2017) Fate of a stressed therapeutic antibody tracked by fluorescence correlation spectroscopy: folded monomers survive aggregation. J Phys Chem B 121:8085–8093
Senga Y, Imamura H, Miyafusa T et al (2017) AlphaScreen-based homogeneous assay using a pair of 25-residue artificial proteins for high-throughput analysis of non-native IgG. Sci Rep 7:12466
den Engelsman J, Garidel P, Smulders R et al (2011) Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm Res 28:920–933
Senga Y, Imamura H, Ogura T et al (2019) In-solution microscopic imaging of fractal aggregates of a stressed therapeutic antibody. Anal Chem 91:4640–4648
Okada T, Ogura T (2016) Nanoscale imaging of untreated mammalian cells in a medium with low radiation damage using scanning electron-assisted dielectric microscopy. Sci Rep 6:29169
Ogura T (2014) Direct observation of unstained biological specimens in water by the frequency transmission electric-field method using SEM. PLoS One 9:e92780
Ogura T (2014) Non-destructive observation of intact bacteria and viruses in water by the highly sensitive frequency transmission electric-field method based on SEM. Biochem Biophys Res Commun 450:1684–1689
Ogura T (2015) Nanoscale analysis of unstained biological specimens in water without radiation damage using high-resolution frequency transmission electric-field system based on FE-SEM. Biochem Biophys Res Commun 459:521–528
Ogura T, Okada T (2017) Nanoscale observation of the natural structure of Milk-fat globules and casein micelles in the liquid condition using a scanning electron assisted dielectric microscopy. Biochem Biophys Res Commun 491:1021–1025
Okada T, Ogura T (2018) Nanoscale imaging of the adhesion core including integrin β1 on intact living cells using scanning electron-assisted dielectric-impedance microscopy. PLoS One 13:e0204133
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Imamura H, Honda S (2019) pH-shift stress on antibodies. Methods Enzymol 622:329–334
Eggersdorfer ML, Kadau D, Herrmann HJ et al (2012) Aggregate morphology evolution by sintering: number and diameter of primary particles. J Aerosol Sci 46:7–19
Martos FJ, Lapuerta M, Expósito JJ et al (2017) Overestimation of the fractal dimension from projections of soot agglomerates. Powder Technol 311:528–536
Halley JM, Hartley S, Kallimanis AS et al (2004) Uses and abuses of fractal methodology in ecology. Ecol Lett 7:254–271
Ogura T, Sato C (2001) An automatic particle pickup method using a neural network applicable to low-contrast electron micrographs. J Struct Biol 136:227–238
Acknowledgments
This study was supported by the Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Scientific Research (B) (JP19H03230 and JP19H03363) and Japan Science and Technology Agency CREST Grant Number JPMJCR19H2, Japan. Yukako Senga and Toshihiko Ogura contributed equally to this work.
Competing financial interests: T.O. is a named inventor on pending patents for the SE-ADM system.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix
Appendix
The following is an example code of the function FractalBoxCounter written in IGOR Pro version 6.22A for fractal analysis using the box-counting method. A threshold (see Subheading 3.4, step 6) and box size values (see Note 19) should be input, and pixels with values below the threshold are regarded as particles (see also related instructions in the following code). Python code is also available (FractalBoxCounter.py). These IGOR Pro and Python codes are compatible with “FractalBoxCounter.java” installed in ImageJ (see Note 21) and allows for modifications depending on the purpose (e.g., quick assessment of various thresholds).
The example code, FractalBoxCounter, for IGOR Pro.
Function FractalBoxCounter() // README (instruction) // 1. Load your image (e.g., img.tif) via "Data --> Load Waves --> Load image..." // 2. Rename the 2D wave of your image data to "woi" via "Data --> Rename..."; or type "rename 'img.tif', woi" in the command line. // 3. (optional) Type "ImageTransform convert2gray woi; woi = M_Image2Gray" in the command line if your image loaded is not 8-bit gray scaled. // 4. Input your values of the threshold (default; 160) and box size values (default; 2,3,4,6,8,12,16,32,64); see User-defined parameters. // 5. Type "FractalBoxCounter()" in the command line. Wave woi // 2D Wave of your Original Image (8-bit gray scale) ///// User-Defined Parameters ///// Variable threshold = 160 // Threshold Make/O/D box_size = {2,3,4,6,8,12,16,32,64} // Box size values (pixel) ///// Variables //////////////////// Variable width = dimsize(woi,0) Variable height = dimsize(woi,1) Variable left, right, top, bottom Variable foreground_count Variable i, j, k ///// Make binary image //////////// Make/O/D/N=(width,height) wbi // 2D Wave of a Binary Image Variable vp // Value of a given Pixel j = 0 do i = 0 do vp = woi[i][j] if (vp > threshold) // 0 (black) < vp < 255 (white) wbi[i][j] = 0 // no object else wbi[i][j] = 1 // object endif i += 1 while (i < width) j += 1 while (j < height) ///// Find Margins ////////////////// //Find left edge left = -1 do left += 1 if (left >= width) print "Error: No non-background pixels found" Abort endif i = 0; foreground_count = 0 do foreground_count += wbi[left][i] i += 1 while (i < height) while (foreground_count == 0) //Find top edge top = -1 do top += 1 i = left; foreground_count = 0 do foreground_count += wbi[i][top] i += 1 while (i < width - left) while (foreground_count == 0) //Find right edge right = width + 1 do right -= 1 i = top; foreground_count = 0 do foreground_count += wbi[right - 1][i] i += 1 while (i < (height - top)) while (foreground_count == 0) //Find bottom edge bottom = height + 1 do bottom -= 1 i = left; foreground_count = 0 do foreground_count += wbi[i][bottom - 1] i += 1 while (i < (right - left)) while (foreground_count == 0) ///// Box Counting ////////////////// Variable n_size = numpnts(box_size) // Number of boxes Make/O/D/N=(n_size) counts, ln_box_size, ln_counts Variable count Variable size // box size Variable x, y Variable w, h // box width, box height k = 0 do count = 0 size = box_size[k] y = top do x = left do // Tuning box sizes // if (size <= (right-left)) w = size if ((x + size) >= right) w = right - x // Reduce box sizes at the edge// endif else w = (right - left) // when user-defined box size > image size endif if (size <= (bottom-top)) h = size if ((y + size) >= bottom) h = bottom - y // Reduce box sizes at the edge// endif else h = (bottom-top) // when user-defined box size > image size endif ///// Counting foreground pixels //////// foreground_count = 0 j = 0 do i = 0 do foreground_count += wbi[x + i][y + j] i += 1 while (i < w) j += 1 while (j < h) if (foreground_count != 0) count+=1 endif /////////////////////////////////////// x += size while (x < right) y += size while (y < bottom) counts[k] = count k += 1 while (k < (n_size)) ln_box_size = ln(box_size) ln_counts = ln(counts) ///// Least square for ln_counts vs. ln_box_size /////// Variable D_2d // 2D fractal dimension Variable avg_x, avg_y, sum_xy, sum_x2 avg_x = 0; avg_y =0; sum_xy = 0; sum_x2 = 0 i = 0 do avg_x += ln_box_size[i]; avg_y += ln_counts[i] i+=1 while (i < n_size) avg_x = avg_x / n_size; avg_y = avg_y / n_size i = 0 do sum_xy += (ln_box_size[i] - avg_x)*(ln_counts[i] - avg_y) sum_x2 += (ln_box_size[i] - avg_x)^2 i+=1 while (i< n_size) D_2d = -1*sum_xy / sum_x2; Print "Fractal dimension: ", D_2d return D_2d end
The example code, FractalBoxCounter.py, for Python
## This program supports python 2 and 3. ## Numpy (the package for scientific computing) and Pillow (Python Imaging Library) are required to run this program. ## Run the following command ## python FractalBoxCounter.py 160 input.tiff output.tiff ## 160 indicates the threshold. Change the value depending on your data. ## input.tiff indicates your input image file. ## output.tiff indicates the file name of output binary image. from PIL import Image import numpy as np import sys ### User-defined parameters ### th = int(sys.argv[1]) # Threshold. box_size_values = [2,3,4,6,8,12,16,32,64] #// Box size values. Integer. ### Load your image ### img = Image.open(sys.argv[2]).convert('L') #sys.argv[2] for your original image. convert('L') for 8-bit gray scaling. imgArray = np.asarray(img) maxcol, maxrow = img.size ### Make binary image ### imgArray_bin = np.zeros((maxrow,maxcol)) # binary image for output wbi = np.zeros((maxrow,maxcol)) # binary image for box counting for i in range(maxrow): for j in range(maxcol): if(imgArray[i,j] > th): # 0 (black) --> 255 (white) imgArray_bin[i,j] = 255 # 255 gives white wbi[i,j] = 0 else: imgArray_bin[i,j] = 0 # zero gives black wbi[i,j] = 1 # Output the binary image imgout = Image.fromarray(np.uint8(imgArray_bin)) imgout.save(sys.argv[3]) #sys.argv[3] for output file name ### Find margins ### width = maxcol height = maxrow foreground_count = 0 left = -1 while foreground_count == 0: left += 1 if (left >= width): print('Error: No non-background pixels found') sys.exit() i = 0 foreground_count = 0 while i < height: foreground_count += wbi[i,left] i += 1 # Find top edge foreground_count = 0 top = -1 while foreground_count == 0: top += 1 i = left foreground_count = 0 while i < (width - left): foreground_count += wbi[top,i] i += 1 # Find right edge foreground_count = 0 right = width + 1 while foreground_count == 0: right -= 1 i = top foreground_count = 0 while i < (height - top): foreground_count += wbi[i,right-1] i += 1 # Find bottom edge foreground_count = 0 bottom = height + 1 while foreground_count == 0: bottom -= 1 i = left foreground_count = 0 while i < (right - left): foreground_count += wbi[bottom-1,i] i += 1 ### Box counting ### n_size = len(box_size_values) #// Number of boxes box_size = np.array(box_size_values) counts = np.zeros(n_size) ln_box_size = np.zeros(n_size) ln_counts = np.zeros(n_size) k = 0 while (k < (n_size)): count = 0 size = box_size[k] y = top while (y < bottom): x = left while (x < right): # Tuning box sizes if (size <= (right-left)): w = size # box width if ((x + size) >= right): w = right - x # Reduce box sizes at the edge else: w = (right - left) # when user-defined box size > image size if (size <= (bottom-top)): h = size # box height if ((y + size) >= bottom): h = bottom - y # Reduce box sizes at the edge else: h = (bottom-top) # when user-defined box size > image size # Counting foreground pixels foreground_count = 0 j = 0 while (j < h): i = 0 while (i < w): foreground_count += wbi[y + j][x + i] i += 1 j += 1 if (foreground_count != 0): count += 1 x += size y += size counts[k] = count k += 1 ### Fractal dimension ### ln_box_size = np.log(box_size) # float ln_counts = np.log(counts) # float # Least square for ln_counts vs. ln_box_size avg_x = np.average(ln_box_size) avg_y = np.average(ln_counts) sum_xy = 0 sum_x2 = 0 i = 0 while (i < n_size): sum_xy += (ln_box_size[i] - avg_x)*(ln_counts[i] - avg_y) sum_x2 += (ln_box_size[i] - avg_x)*(ln_box_size[i] - avg_x) i += 1 D_2d = -1*sum_xy / sum_x2 # 2D fractal dimension print('Fractal dimension = ',D_2d)
Rights and permissions
Copyright information
© 2022 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Senga, Y., Ogura, T., Imamura, H., Honda, S. (2022). Nano-Microscopy of Therapeutic Antibody Aggregates in Solution. In: Houen, G. (eds) Therapeutic Antibodies. Methods in Molecular Biology, vol 2313. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1450-1_13
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1450-1_13
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1449-5
Online ISBN: 978-1-0716-1450-1
eBook Packages: Springer Protocols