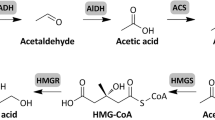
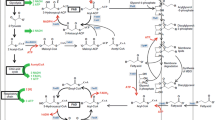
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Vickers C, Klein-Marcuschamer D, Krömer J (2012) Examining the feasibility of bulk commodity production in Escherichia coli. Biotechnol Lett 34:585–596
Zeng AP, Biebl H (2002) Bulk chemicals from biotechnology: the case of 1,3-propanediol production and the new trends. Adv Biochem Eng Biotechnol 74:239–259
Chen X, Zhou L, Tian K et al (2013) Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol Adv 31:1200–1223
Keasling JD (2010) Manufacturing molecules through metabolic engineering. Science 330:1355–1358
Taher H, Al-Zuhair S, Al-Marzouqi AH et al (2011) A review of enzymatic transesterification of microalgal oil-based biodiesel using supercritical technology. Enzyme Res 2011:468292
U.S. Department of Energy EIA (2012) Petroleum marketing monthly. Department of Energy, Washington, DC, DOE Publ. No. EIA-0380(20012/02)
U.S. Department of Energy EIA (2006) Annual energy outlook 2006 with projections to 2030. Department of Energy, DOE Publ. No. EIA-0383(2006), Washington, DC
U.S. Department of Energy EIA (2007) Annual energy outlook 2007 with projections to 2030. Departmentt of Energy, DOE/EIA-0383(2007), Washington, DC
Lee JW, Na D, Park JM et al (2012) Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol 8:536–546
Demirbas A (2009) Political, economic and environmental impacts of biofuels: a review. 86. Appl Energy 86(1):S108–S117
Jang YS, Kim B, Shin JH et al (2012) Bio-based production of C2-C6 platform chemicals. Biotechnol Bioeng 109:2437–2459
Yim H, Haselbeck R, Niu W et al (2011) Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 7:445–452
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89
Jambunathan P, Zhang K (2014) Novel pathways and products from 2-keto acids. Curr Opin Biotechnol 29:1–7
Schiweck H, Bär A, Vogel R et al (2000) Sugar Alcohols, in Ullmann’s Encyclopedia of Industrial Chemistry. Wiley-VCH Verlag GmbH & Co, KGaA
Shin JH, Kim HU, Kim DI et al (2013) Production of bulk chemicals via novel metabolic pathways in microorganisms. Biotechnol Adv 31:925–935
Jarboe LR, Liu P, Kautharapu KB et al (2012) Optimization of enzyme parameters for fermentative production of biorenewable fuels and chemicals. Comput Struct Biotechnol J 3:e201210005
Stephanopoulos G (2012) Synthetic biology and metabolic engineering. ACS Synth Biol 1:514–525
Nielsen J, Fussenegger M, Keasling J et al (2014) Engineering synergy in biotechnology. Nat Chem Biol 10:319–322
Schirmer A, Rude MA, Li X et al (2010) Microbial biosynthesis of alkanes. Science 329:559–562
Yu C, Cao Y, Zou H et al (2011) Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols. Appl Microbiol Biotechnol 89:573–583
Zhou L, Tian KM, Niu DD et al (2012) Improvement of D-lactate productivity in recombinant Escherichia coli by coupling production with growth. Biotechnol Lett 34:1123–1130
Zhou L, Niu DD, Tian KM et al (2012) Genetically switched D-lactate production in Escherichia coli. Metab Eng 14:560–568
Utrilla J, Licona-Cassani C, Marcellin E et al (2012) Engineering and adaptive evolution of Escherichia coli for D-lactate fermentation reveals GatC as a xylose transporter. Metab Eng 14:469–476
Zhou L, Zuo ZR, Chen XZ et al (2011) Evaluation of genetic manipulation strategies on D-lactate production by Escherichia coli. Curr Microbiol 62:981–989
Yang J, Wang Z, Zhu N et al (2014) Metabolic engineering of Escherichia coli and in silico comparing of carboxylation pathways for high succinate productivity under aerobic conditions. Microbiol Res 169:432–440
Jiang M, Chen X, Liang L et al (2014) Co-expression of phosphoenolpyruvate carboxykinase and nicotinic acid phosphoribosyltransferase for succinate production in engineered Escherichia coli. Enzyme Microb Technol 56:8–14
Tang J, Zhu X, Lu J et al (2013) Recruiting alternative glucose utilization pathways for improving succinate production. Appl Microbiol Biotechnol 97:2513–2520
Tan Z, Zhu X, Chen J et al (2013) Activating phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in combination for improvement of succinate production. Appl Environ Microbiol 79:4838–4844
Kim K, Kim SK, Park YC et al (2014) Enhanced production of 3-hydroxypropionic acid from glycerol by modulation of glycerol metabolism in recombinant Escherichia coli. Bioresour Technol 156:170–175
Jung WS, Kang JH, Chu HS et al (2014) Elevated production of 3-hydroxypropionic acid by metabolic engineering of the glycerol metabolism in Escherichia coli. Metab Eng 23:116–122
Kwak S, Park YC, Seo JH (2013) Biosynthesis of 3-hydroxypropionic acid from glycerol in recombinant Escherichia coli expressing Lactobacillus brevis dhaB and dhaR gene clusters and E. coli K-12 aldH. Bioresour Technol 135:432–439
Rathnasingh C, Raj SM, Lee Y et al (2012) Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J Biotechnol 157:633–640
Desai SH, Rabinovitch-Deere CA, Tashiro Y et al (2014) Isobutanol production from cellobiose in Escherichia coli. Appl Microbiol Biotechnol 98:3727–3736
Shi A, Zhu X, Lu J et al (2013) Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab Eng 16:1–10
Trinh CT, Li J, Blanch HW et al (2011) Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl Environ Microbiol 77:4894–4904
Hwang HJ, Park JH, Kim JH et al (2014) Engineering of a butyraldehyde dehydrogenase of Clostridium saccharoperbutylacetonicum to fit an engineered 1,4-butanediol pathway in Escherichia coli. Biotechnol Bioeng 111:1374–1384
Wang W, Lu X (2013) Microbial synthesis of alka(e)nes. Front Bioeng Biotechnol. doi: 10.3389/fbioe.2013.00010
Howard TP, Middelhaufe S, Moore K et al (2013) Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proc Natl Acad Sci U S A 110:7636–7641
Riemer SA, Rex R, Schomburg D (2013) A metabolite-centric view on flux distributions in genome-scale metabolic models. BMC Syst Biol 7:33
McCloskey D, Palsson BO, Feist AM (2013) Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli. Mol Syst Biol 9:661
Xu Z, Sun X, Sun J (2013) Construction and analysis of the model of energy metabolism in E. coli. PLoS One 8:e55137
Khodayari A, Zomorrodi AR, Liao JC et al (2014) A kinetic model of Escherichia coli core metabolism satisfying multiple sets of mutant flux data. Metab Eng. doi:10.1016/j.ymben.2014.05.014
Zomorrodi AR, Lafontaine Rivera JG, Liao JC et al (2013) Optimization-driven identification of genetic perturbations accelerates the convergence of model parameters in ensemble modeling of metabolic networks. Biotechnol J 8:1090–1104
Xu Z, Zheng P, Sun J et al (2013) ReacKnock: identifying reaction deletion strategies for microbial strain optimization based on genome-scale metabolic network. PLoS One 8:e72150
Chang RL, Andrews K, Kim D et al (2013) Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli. Science 340:1220–1223
Durot M, Bourguignon P-Y, Schachter V (2009) Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev 33:164–190
O’Brien EJ, Lerman JA, Chang RL et al (2013) Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol Syst Biol 9:693
Cho BK, Kim D, Knight EM et al (2014) Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol 12:4
Atsumi S, Liao JC (2008) Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol 19:414–419
Marcheschi RJ, Li H, Zhang K et al (2012) A synthetic recursive “+1” pathway for carbon chain elongation. ACS Chem Biol 7:689–697
Planson AG, Carbonell P, Grigoras I et al (2012) A retrosynthetic biology approach to therapeutics: from conception to delivery. Curr Opin Biotechnol 23:948–956
Rodrigo G, Carrera J, Prather KJ et al (2008) DESHARKY: automatic design of metabolic pathways for optimal cell growth. Bioinformatics 24:2554–2556
Hatzimanikatis V, Li C, Ionita JA et al (2005) Exploring the diversity of complex metabolic networks. Bioinformatics 21:1603–1609
Cho A, Yun H, Park JH et al (2010) Prediction of novel synthetic pathways for the production of desired chemicals. BMC Syst Biol 4:35
Mavrovouniotis M, Stephanopoulos G (1992) Synthesis of biochemical production routes. Comput Chem Eng 16:605–619
Tian J, Gong H, Sheng N et al (2004) Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432:1050–1054
Quan J, Saaem I, Tang N et al (2011) Parallel on-chip gene synthesis and application to optimization of protein expression. Nat Biotechnol 29:449–452
Shetty RP, Endy D, Knight TF Jr (2008) Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2:5
Xu P, Vansiri A, Bhan N et al (2012) ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth Biol 1:256–266
Ellis T, Adie T, Baldwin GS (2011) DNA assembly for synthetic biology: from parts to pathways and beyond. Integr Biol (Camb) 3:109–118
Hillson N (2011) DNA Assembly method standardization for synthetic biomolecular circuits and systems. In: Koeppl H, Setti G, di Bernardo M et al (eds) Design and analysis of biomolecular circuits. Springer, New York, pp 295–314
Quan J, Tian J (2014) Circular polymerase extension cloning. Methods Mol Biol 1116:103–117
Li MZ, Elledge SJ (2007) Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4:251–256
Gibson DG, Young L, Chuang RY et al (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345
Shao Z, Zhao H (2009) DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res 37:e16
Du J, Yuan Y, Si T et al (2012) Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res 40:e142
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Jarboe LR, Zhang X, Wang X et al (2010) Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology. J Biomed Biotechnol 2010:761042
Gaj T, Sirk SJ, Barbas CF (2014) Expanding the scope of site-specific recombinases for genetic and metabolic engineering. Biotechnol Bioeng 111:1–15
Posfai G, Koob MD, Kirkpatrick HA et al (1997) Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol 179:4426–4428
Jantama K, Zhang X, Moore JC et al (2008) Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng 101:881–893
Zhang X, Jantama K, Moore JC et al (2007) Production of L -alanine by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 77:355–366
Gay P, Le Coq D, Steinmetz M et al (1985) Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol 164:918–921
Hammer K, Mijakovic I, Jensen PR (2006) Synthetic promoter libraries–tuning of gene expression. Trends Biotechnol 24:53–55
Nevoigt E, Kohnke J, Fischer CR et al (2006) Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl Environ Microbiol 72:5266–5273
Alper H, Fischer C, Nevoigt E et al (2005) Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA 102:12678–12683
Solem C, Jensen PR (2002) Modulation of gene expression made easy. Appl Environ Microbiol 68:2397–2403
Jensen PR, Hammer K (1998) The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol 64:82–87
Jensen PR, Hammer K (1998) Artificial promoters for metabolic optimization. Biotechnol Bioeng 58:191–195
de la Cueva-Mendez G, Pimentel B (2007) Gene and cell survival: lessons from prokaryotic plasmid R1. EMBO Rep 8:458–464
Keasling JD (2008) Synthetic biology for synthetic chemistry. ACS Chem Biol 3:64–76
Lu J, Tang J, Liu Y et al (2012) Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl Microbiol Biotechnol 93:2455–2462
De Mey M, Maertens J, Boogmans S et al (2010) Promoter knock-in: a novel rational method for the fine tuning of genes. BMC Biotechnol 10:26
Meynial-Salles I, Cervin MA, Soucaille P (2005) New tool for metabolic pathway engineering in Escherichia coli: one-step method to modulate expression of chromosomal genes. Appl Environ Microbiol 71:2140–2144
Millard CS, Chao YP, Liao JC et al (1996) Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl Environ Microbiol 62:1808–1810
Santos CN, Stephanopoulos G (2008) Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol 12:168–176
Pfleger BF, Pitera DJ, Smolke CD et al (2006) Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol 24:1027–1032
Dueber JE, Wu GC, Malmirchegini GR et al (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27:753–759
Cobb RE, Sun N, Zhao H (2013) Directed evolution as a powerful synthetic biology tool. Methods 60:81–90
Wang HH, Isaacs FJ, Carr PA et al (2009) Programming cells by multiplex genome engineering and accelerated evolution. Nature 460:894–898
Yoo SM, Na D, Lee SY (2013) Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nat Protoc 8:1694–1707
Xu P, Wang W, Li L et al (2014) Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli. ACS Chem Biol 9:451–458
Farmer WR, Liao JC (2000) Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol 18:533–537
Zhang F, Carothers JM, Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol 30:354–359
Cobb RE, Si T, Zhao H (2012) Directed evolution: an evolving and enabling synthetic biology tool. Curr Opin Chem Biol 16:285–291
Zhou S, Yomano LP, Shanmugam KT et al (2005) Fermentation of 10 % (w/v) sugar to D: (-)-lactate by engineered Escherichia coli B. Biotechnol Lett 27:1891–1896
Zhou S, Shanmugam KT, Yomano LP et al (2006) Fermentation of 12 % (w/v) glucose to 1.2 M lactate by Escherichia coli strain SZ194 using mineral salts medium. Biotechnol Lett 28:663–670
Grabar TB, Zhou S, Shanmugam KT et al (2006) Methylglyoxal bypass identified as source of chiral contamination in l(+) and d(-)-lactate fermentations by recombinant Escherichia coli. Biotechnol Lett 28:1527–1535
Zhang X, Shanmugam KT, Ingram LO (2010) Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol 76:2397–2401
Zhang X, Jantama K, Moore JC et al (2009) Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc Natl Acad Sci USA 106:20180–20185
Jiang M, Wan Q, Liu R et al (2014) Succinic acid production from corn stalk hydrolysate in an E. coli mutant generated by atmospheric and room-temperature plasmas and metabolic evolution strategies. J Ind Microbiol Biotechnol 41:115–123
Trinh CT, Srienc F (2009) Metabolic engineering of Escherichia coli for efficient conversion of glycerol to ethanol. Appl Environ Microbiol 75:6696–6705
Fernandez-Sandoval MT, Huerta-Beristain G, Trujillo-Martinez B et al (2012) Laboratory metabolic evolution improves acetate tolerance and growth on acetate of ethanologenic Escherichia coli under non-aerated conditions in glucose-mineral medium. Appl Microbiol Biotechnol 96:1291–1300
Zheng H, Wang X, Yomano LP et al (2013) Improving Escherichia coli FucO for furfural tolerance by saturation mutagenesis of individual amino acid positions. Appl Environ Microbiol 79:3202–3208
Liu H, Yan M, Lai C et al (2010) gTME for improved xylose fermentation of Saccharomyces cerevisiae. Appl Biochem Biotechnol 160:574–582
Tyo KE, Alper HS, Stephanopoulos GN (2007) Expanding the metabolic engineering toolbox: more options to engineer cells. Trends Biotechnol 25:132–137
Alper H, Stephanopoulos G (2007) Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng 9:258–267
Alper H, Moxley J, Nevoigt E et al (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565–1568
Shi DJ, Wang CL, Wang KM (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36:139–147
Otte B, Grunwaldt E, Mahmoud O et al (2009) Genome shuffling in Clostridium diolis DSM 15410 for improved 1,3-propanediol production. Appl Environ Microbiol 75:7610–7616
Hida H, Yamada T, Yamada Y (2007) Genome shuffling of Streptomyces sp. U121 for improved production of hydroxycitric acid. Appl Microbiol Biotechnol 73:1387–1393
Zhang YX, Perry K, Vinci VA et al (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Warner JR, Reeder PJ, Karimpour-Fard A et al (2010) Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol 28:856–862
Royce LA, Boggess E, Fu Y et al (2014) Transcriptomic analysis of carboxylic acid challenge in Escherichia coli: beyond membrane damage. PLoS One 9:e89580
McCloskey D, Gangoiti JA, King ZA et al (2014) A model-driven quantitative metabolomics analysis of aerobic and anaerobic metabolism in E. coli K-12 MG1655 that is biochemically and thermodynamically consistent. Biotechnol Bioeng 111:803–815
Wiback SJ, Mahadevan R, Palsson BO (2004) Using metabolic flux data to further constrain the metabolic solution space and predict internal flux patterns: the Escherichia coli spectrum. Biotechnol Bioeng 86:317–331
Oliver DJ, Nikolau B, Wurtele ES (2002) Functional genomics: high-throughput mRNA, protein, and metabolite analyses. Metab Eng 4:98–106
Patterson SD, Aebersold RH (2003) Proteomics: the first decade and beyond. Nat Genet 33(Suppl):311–323
Alonso S, Rendueles M, Diaz M (2014) Microbial production of specialty organic acids from renewable and waste materials. Crit Rev Biotechnol . doi:10.3109/07388551.2014.904269
Tsao GT, Cao NJ, Du J et al (1999) Production of multifunctional organic acids from renewable resources. In: Tsao GT, Brainard AP, Bungay HR et al (eds) Recent progress in bioconversion of Lignocellulosics. Springer, Berlin, pp 243–280
Zhou S, Causey TB, Hasona A et al (2003) Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol 69:399–407
Shukla VB, Zhou S, Yomano LP et al (2004) Production of D(-)-lactate from sucrose and molasses. Biotechnol Lett 26:689–693
Wang Q, Yang P, Liu C et al (2013) Biosynthesis of poly(3-hydroxypropionate) from glycerol by recombinant Escherichia coli. Bioresour Technol 131:548–551
Meng DC, Shi ZY, Wu LP et al (2012) Production and characterization of poly(3-hydroxypropionate-co-4-hydroxybutyrate) with fully controllable structures by recombinant Escherichia coli containing an engineered pathway. Metab Eng 14:317–324
Werpy T GPe (2004) Top value added chemicals from biomass. U.S. Department of Energy, Washington, DC. http://www1.eere.energy.gov/biomass/pdfs/35523.pdf
Holo H (1989) Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch Microbiol 151:252–256
Strauss G, Eisenreich W, Bacher A et al (1992) 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing Archaebacterium Thermoproteus neutrophilus and in the phototrophic Eubacterium Chloroflexus aurantiacus. Eur J Biochem 205:853–866
Hugler M, Huber H, Stetter KO et al (2003) Autotrophic CO2 fixation pathways in archaea (Crenarchaeota). Arch Microbiol 179:160–173
Berg IA, Kockelkorn D, Buckel W et al (2007) A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786
Ansede JH, Pellechia PJ, Yoch DC (1999) Metabolism of acrylate to beta-hydroxypropionate and its role in dimethylsulfoniopropionate lyase induction by a salt marsh sediment bacterium, Alcaligenes faecalis M3A. Appl Environ Microbiol 65:5075–5081
Loh KD, Gyaneshwar P, Papadimitriou EM et al (2006) A previously undescribed pathway for pyrimidine catabolism. Proc Natl Acad Sci U S A 103:5114–5119
Kim KS, Pelton JG, Inwood WB et al (2010) The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J Bacteriol 192:4089–4102
Andersen G, Bjornberg O, Polakova S et al (2008) A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J Mol Biol 380:656–666
Valdehuesa KN, Liu H, Nisola GM et al (2013) Recent advances in the metabolic engineering of microorganisms for the production of 3-hydroxypropionic acid as C3 platform chemical. Appl Microbiol Biotechnol 97:3309–3321
Kumar V, Ashok S, Park S (2013) Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol Adv 31:945–961
Jiang X, Meng X, Xian M (2009) Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol 82:995–1003
Raj S, Rathnasingh C, Jo J-E et al (2008) Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem 43:1440–1446
Mohan Raj S, Rathnasingh C, Jung WC et al (2009) Effect of process parameters on 3-hydroxypropionic acid production from glycerol using a recombinant Escherichia coli. Appl Microbiol Biotechnol 84:649–657
Rathnasingh C, Raj SM, Jo JE et al (2009) Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng 104:729–739
Tokuyama K, Ohno S, Yoshikawa K et al (2014) Increased 3-hydroxypropionic acid production from glycerol, by modification of central metabolism in Escherichia coli. Microb Cell Fact 13:64
Bunch PK, Mat-Jan F, Lee N et al (1997) The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143(Pt 1):187–195
Chatterjee R, Millard CS, Champion K et al (2001) Mutation of the ptsG gene results in increased production of succinate in fermentation of glucose by Escherichia coli. Appl Environ Microbiol 67:148–154
Donnelly MI, Millard CS, Clark DP et al (1998) A novel fermentation pathway in an Escherichia coli mutant producing succinic acid, acetic acid, and ethanol. Appl Biochem Biotechnol 70–72:187–198
Stols L, Donnelly MI (1997) Production of succinic acid through overexpression of NAD(+)-dependent malic enzyme in an Escherichia coli mutant. Appl Environ Microbiol 63:2695–2701
Escalante A, Cervantes AS, Gosset G et al (2012) Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: peculiarities of regulation and impact on growth and product formation. Appl Microbiol Biotechnol 94:1483–1494
Gabor E, Gohler AK, Kosfeld A et al (2011) The phosphoenolpyruvate-dependent glucose-phosphotransferase system from Escherichia coli K-12 as the center of a network regulating carbohydrate flux in the cell. Eur J Cell Biol 90:711–720
Postma PW, Lengeler JW, Jacobson GR (1993) Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57:543–594
Vemuri GN, Eiteman MA, Altman E (2002) Succinate production in dual-phase Escherichia coli fermentations depends on the time of transition from aerobic to anaerobic conditions. J Ind Microbiol Biotechnol 28:325–332
Sanchez AM, Bennett GN, San KY (2005) Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab Eng 7:229–239
Cheng KK, Wang GY, Zeng J et al (2013) Improved succinate production by metabolic engineering. Biomed Res Int 2013:538790
Clark DP (1989) The fermentation pathways of Escherichia coli. FEMS Microbiol Rev 5:223–234
Shanmugam KT, Ingram LO (2008) Engineering biocatalysts for production of commodity chemicals. J Mol Microbiol Biotechnol 15:8–15
Maloy SR, Nunn WD (1982) Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J Bacteriol 149:173–180
Lorca GL, Ezersky A, Lunin VV et al (2007) Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J Biol Chem 282:16476–16491
Yamamoto K, Ishihama A (2003) Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mol Microbiol 47:183–194
Cozzone AJ, El-Mansi M (2005) Control of isocitrate dehydrogenase catalytic activity by protein phosphorylation in Escherichia coli. J Mol Microbiol Biotechnol 9:132–146
Balzer GJ, Thakker C, Bennett GN et al (2013) Metabolic engineering of Escherichia coli to minimize byproduct formate and improving succinate productivity through increasing NADH availability by heterologous expression of NAD(+)-dependent formate dehydrogenase. Metab Eng 20:1–8
Stols L, Kulkarni G, Harris BG et al (1997) Expression of Ascaris suum malic enzyme in a mutant Escherichia coli allows production of succinic acid from glucose. Appl Biochem Biotechnol 63–65:153–158
Jantama K, Haupt MJ, Svoronos SA et al (2008) Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng 99:1140–1153
Gosset G (2005) Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb Cell Fact 4:14
Zhang X, Jantama K, Shanmugam KT et al (2009) Reengineering Escherichia coli for Succinate Production in Mineral Salts Medium. Appl Environ Microbiol 75:7807–7813
Zhu X, Tan Z, Xu H et al (2014) Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli. Metab Eng 24:87–96
Steiert PS, Stauffer LT, Stauffer GV (1990) The lpd gene product functions as the L protein in the Escherichia coli glycine cleavage enzyme system. J Bacteriol 172:6142–6144
Guest JR, Creaghan IT (1974) Further studies with lipoamide dehydrogenase mutants of Escherichia coli K12. J Gen Microbiol 81:237–245
Guest JR, Creaghan IT (1972) Lipoamide dehydrogenase mutants of Escherichia coli K 12. Biochem J 130:8
Kim YM, Cho HS, Jung GY et al (2011) Engineering the pentose phosphate pathway to improve hydrogen yield in recombinant Escherichia coli. Biotechnol Bioeng 108:2941–2946
Sauer U, Canonaco F, Heri S et al (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619
Sauer U, Lasko DR, Fiaux J et al (1999) Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol 181:6679–6688
Battat E, Peleg Y, Bercovitz A et al (1991) Optimization of L-malic acid production by Aspergillus flavus in a stirred fermentor. Biotechnol Bioeng 37:1108–1116
Peleg Y, Rokem JS, Goldberg I (1990) A simple plate-assay for the screening of L-malic acid producing microorganisms. FEMS Microbiol Lett 55:233–236
Pines O, Even-Ram S, Elnathan N et al (1996) The cytosolic pathway of L-malic acid synthesis in Saccharomyces cerevisiae: the role of fumarase. Appl Microbiol Biotechnol 46:393–399
Taing O, Taing K (2007) Production of malic and succinic acids by sugar-tolerant yeast Zygosaccharomyces rouxii. Eur Food Res Technol 224:343–347
Kawagoe M, Hyakumura K, Suye S-I et al (1997) Application of bubble column fermentors to submerged culture of Schizophyllum commune for production of L-malic acid. J Ferment Bioeng 84:333–336
Zhang X, Wang X, Shanmugam KT et al (2011) L-malate production by metabolically engineered Escherichia coli. Appl Environ Microbiol 77:427–434
Straathof AJ, van Gulik WM (2012) Production of fumaric Acid by fermentation. Subcell Biochem 64:225–240
Ehrlich F (1911) Über die bildung von fumarsäure durch schimmelpilze. Ber Dtsch Chem Ges 44:3737–3742
Wang G, Huang D, Qi H et al (2013) Rational medium optimization based on comparative metabolic profiling analysis to improve fumaric acid production. Bioresour Technol 137:1–8
Zhou Y, Nie K, Zhang X et al (2014) Production of fumaric acid from biodiesel-derived crude glycerol by Rhizopus arrhizus. Bioresour Technol 163:48–53
Gu S, Xu Q, Huang H et al (2014) Alternative respiration and fumaric acid production of Rhizopus oryzae. Appl Microbiol Biotechnol 98:5145–5152
Goldberg I, Steiglitz B (1985) Improved rate of fumaric acid production by Tweens and vegetable oils in rhizopus arrhizus. Biotechnol Bioeng 27:1067–1069
Zhou Y, Du J, Tsao GT (2002) Comparison of fumaric acid production by Rhizopus oryzae using different neutralizing agents. Bioprocess Biosyst Eng 25:179–181
Fu YQ, Li S, Chen Y et al (2010) Enhancement of fumaric acid production by Rhizopus oryzae using a two-stage dissolved oxygen control strategy. Appl Biochem Biotechnol 162:1031–1038
Xu Q, Li S, Fu Y et al (2010) Two-stage utilization of corn straw by Rhizopus oryzae for fumaric acid production. Bioresour Technol 101:6262–6264
Ding Y, Li S, Dou C et al (2011) Production of fumaric acid by Rhizopus oryzae: role of carbon-nitrogen ratio. Appl Biochem Biotechnol 164:1461–1467
Roa Engel CA, van Gulik WM, Marang L et al (2011) Development of a low pH fermentation strategy for fumaric acid production by Rhizopus oryzae. Enzyme Microb Technol 48:39–47
Zhang B, Skory CD, Yang ST (2012) Metabolic engineering of Rhizopus oryzae: effects of overexpressing pyc and pepc genes on fumaric acid biosynthesis from glucose. Metab Eng 14:512–520
Gu C, Zhou Y, Liu L et al (2013) Production of fumaric acid by immobilized Rhizopus arrhizus on net. Bioresour Technol 131:303–307
Ling LB, Ng TK (1989). US Patent 4,877,731. Google Patents
Song CW, Kim DI, Choi S et al (2013) Metabolic engineering of Escherichia coli for the production of fumaric acid. Biotechnol Bioeng 110:2025–2034
Singh J, Gupta KP (2003) Calcium glucarate prevents tumor formation in mouse skin. Biomed Environ Sci 16:9–16
Singh J, Gupta KP (2007) Induction of apoptosis by calcium D-glucarate in 7,12-dimethyl benz [a] anthracene-exposed mouse skin. J Environ Pathol Toxicol Oncol 26:63–73
Walaszek Z, Szemraj J, Hanausek M et al (1996) d-Glucaric acid content of various fruits and vegetables and cholesterol-lowering effects of dietary d-glucarate in the rat. Nutr Res 16:673–681
Moon TS, Yoon SH, Lanza AM et al (2009) Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl Environ Microbiol 75:589–595
Moon TS, Dueber JE, Shiue E et al (2010) Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab Eng 12:298–305
Shiue E, Prather KL (2014) Improving D-glucaric acid production from myo-inositol in E. coli by increasing MIOX stability and myo-inositol transport. Metab Eng 22:22–31
Xie NZ, Liang H, Huang RB et al (2014) Biotechnological production of muconic acid: current status and future prospects. Biotechnol Adv 32:615–622
Lin Y, Sun X, Yuan Q et al (2014) Extending shikimate pathway for the production of muconic acid and its precursor salicylic acid in Escherichia coli. Metab Eng 23:62–69
Sun X, Lin Y, Huang Q et al (2013) A novel muconic acid biosynthesis approach by shunting tryptophan biosynthesis via anthranilate. Appl Environ Microbiol 79:4024–4030
Draths KM, Frost JW (1994) Environmentally Compatible Synthesis of Adipic Acid from D-Glucose. J Am Chem Soc 116:399–400
Niu W, Draths KM, Frost JW (2002) Benzene-free synthesis of adipic acid. Biotechnol Prog 18:201–211
Bui V, Lau MK, MacRae D et al (2013) Methods for producing isomers of muconic acid and muconate salts, United States Patent application. US 2013/0030215 A1, Google Patents
Polen T, Spelberg M, Bott M (2013) Toward biotechnological production of adipic acid and precursors from biorenewables. J Biotechnol 167:75–84
Musser MT (2000) Adipic Acid, in Ullmann’s Encyclopedia of Industrial Chemistry. Wiley-VCH Verlag GmbH & Co, KGaA
Yu JL, Xia XX, Zhong JJ et al (2014) Direct biosynthesis of adipic acid from a synthetic pathway in recombinant Escherichia coli. Biotechnol Bioeng. doi:10.1002/bit.25293
Han L, Chen W, Yuan F et al (2013) Biosynthesis of adipic acid. Sheng Wu Gong Cheng Xue Bao 29:1374–1385
Picataggio S, Beardslee T (2012) Biological methods for preparing adipic acid. Google Patents US 8,241,879 B2
Parthasarathy A, Pierik AJ, Kahnt J et al (2011) Substrate specificity of 2-hydroxyglutaryl-CoA dehydratase from Clostridium symbiosum: toward a bio-based production of adipic acid. Biochemistry 50:3540–3550
Noack H, Georgiev V, Blomberg MR et al (2011) Theoretical insights into heme-catalyzed oxidation of cyclohexane to adipic acid. Inorg Chem 50:1194–1202
Burgard AP, Pharkya P, Osterhout RE (2010) Microorganisms for the production of adipic acid and other compounds. Google Patents US 2010/0330626 A1
Dugal M, Sankar G, Raja R et al (2000) Designing a Heterogeneous Catalyst for the Production of Adipic Acid by Aerial Oxidation of Cyclohexane. Angew Chem Int Ed Engl 39:2310–2313
Nakamura CE, Whited GM (2003) Metabolic engineering for the microbial production of 1,3-propanediol. Curr Opin Biotechnol 14:454–459
Jain R, Yan Y (2011) Dehydratase mediated 1-propanol production in metabolically engineered Escherichia coli. Microb Cell Fact 10:97
Altaras NE, Cameron DC (1999) Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli. Appl Environ Microbiol 65:1180–1185
Altaras NE, Cameron DC (2000) Enhanced production of (R)-1,2-propanediol by metabolically engineered Escherichia coli. Biotechnol Prog 16:940–946
Soma Y, Inokuma K, Tanaka T et al (2012) Direct isopropanol production from cellobiose by engineered Escherichia coli using a synthetic pathway and a cell surface display system. J Biosci Bioeng 114:80–85
Lan EI, Liao JC (2013) Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bioresour Technol 135:339–349
Zhang K, Sawaya MR, Eisenberg DS et al (2008) Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci USA 105:20653–20658
Dhande YK, Xiong M, Zhang K (2012) Production of C5 carboxylic acids in engineered Escherichia coli. Process Biochem 47:1965–1971
Bhan N, Xu P, Koffas MA (2013) Pathway and protein engineering approaches to produce novel and commodity small molecules. Curr Opin Biotechnol 24:1137–1143
Baez A, Cho KM, Liao JC (2011) High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl Microbiol Biotechnol 90:1681–1690
Bastian S, Liu X, Meyerowitz JT et al (2011) Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab Eng 13:345–352
Machado HB, Dekishima Y, Luo H et al (2012) A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab Eng 14:504–511
Tseng HC, Prather KL (2012) Controlled biosynthesis of odd-chain fuels and chemicals via engineered modular metabolic pathways. Proc Natl Acad Sci USA 109:17925–17930
Yu P, Tai Y-S, Woodruff AP et al (2012) Engineering artificial metabolic pathways for biosynthesis. Curr Opin Chem Eng 1:373–379
Pitera DJ, Paddon CJ, Newman JD et al (2007) Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng 9:193–207
Chowdhury R, Sahu G, Das J (1996) Stress response in pathogenic bacteria. J Bioscience 21:149–160
Acknowledgments
This research was supported by grants from Tianjin Key Technology R&D program of Tianjin Municipal Science and Technology Commission (11ZCZDSY09100, 13ZCZDSY05300), the National Natural Science Foundation of China (31370136), and the Key Deployment Project of the Chinese Academy of Sciences (KGZD-EW-606; KSZD-EW-Z-016-2).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Liu, P., Zhu, X., Tan, Z., Zhang, X., Ma, Y. (2015). Construction of Escherichia Coli Cell Factories for Production of Organic Acids and Alcohols. In: Ye, Q., Bao, J., Zhong, JJ. (eds) Bioreactor Engineering Research and Industrial Applications I. Advances in Biochemical Engineering/Biotechnology, vol 155. Springer, Berlin, Heidelberg. https://doi.org/10.1007/10_2014_294
Download citation
DOI: https://doi.org/10.1007/10_2014_294
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-49159-1
Online ISBN: 978-3-662-49161-4
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)