Abstract
Several chronic neurodegenerative disorders manifest deposits of misfolded or aggregated proteins. Genetic mutations are the root cause for protein misfolding in rare families, but the majority of patients have sporadic forms possibly related to environmental factors. In some cases, the ubiquitin-proteasome system or molecular chaperones can prevent accumulation of aberrantly folded proteins. Recent studies suggest that generation of excessive nitric oxide (NO) and reactive oxygen species (ROS), in part due to overactivity of the NMDA-subtype of glutamate receptor, can mediate protein misfolding in the absence of genetic predisposition. S-Nitrosylation, or covalent reaction of NO with specific protein thiol groups, represents one mechanism contributing to NO-induced protein misfolding and neurotoxicity. Here, we present evidence suggesting that NO contributes to protein misfolding via S-nitrosylating protein-disulfide isomerase or the E3 ubiquitin ligase parkin. We discuss how memantine/NitroMemantine can inhibit excessive NMDA receptor activity to ameliorate NO production, protein misfolding, and neurodegeneration.
Similar content being viewed by others
Introduction
Many neurodegenerative diseases are characterized by the accumulation of misfolded proteins that adversely affect neuronal connectivity and plasticity, and trigger cell death signaling pathways [1, 2]. For example, degenerating brain contains aberrant accumulations of misfolded, aggregated proteins, such as α-synuclein and synphilin-1 in Parkinson’s disease (PD), and amyloid-β (Aβ) and tau in Alzheimer’s disease (AD). The inclusions observed in PD are called Lewy bodies and are mostly found in the cytoplasm. AD brains show intracellular neurofibrillary tangles, which contain hyperphosphorylated tau, and extracellular plaques, which contain Aβ. These aggregates may consist of oligomeric complexes of non-native secondary structures, and demonstrate poor solubility in aqueous or detergent solvent. Other disorders manifesting protein aggregation include Huntington’s disease (a polyQ disorder), amyotrophic lateral sclerosis (ALS), and prion disease [3]. The aforementioned disorders are also termed “conformational diseases” because of the emergence of protein aggregation in the brain [4].
An additional feature of most neurodegenerative diseases is excessive generation of reactive nitrogen species (RNS) and reactive oxygen species (ROS), which can contribute to neuronal cell injury and death [5–9]. While many intra- and extra-cellular molecules may participate in neuronal injury, accumulation of nitrosative stress due to excessive generation of nitric oxide (NO) appears to be a potential factor contributing to neuronal cell damage and death [10, 11]. A well-established model for NO production entails a central role of the N-methyl-d-aspartate (NMDA)-type glutamate receptors in nervous system. Excessive activation of NMDA receptors drives Ca2+ influx, which in turn activates neuronal NO synthase (nNOS) as well as the generation of ROS [12, 13]. Accumulating evidence suggests that NO can mediate both protective and neurotoxic effects by reacting with cysteine residues of target proteins to form S-nitrosothiols (SNOs), a process termed S-nitrosylation because of its effects on the chemical biology of protein function. Importantly, normal mitochondrial respiration may also generate free radicals, principally ROS, and one such molecule, superoxide anion (O2 −), reacts rapidly with free radical NO to form the very toxic product peroxynitrite (ONOO−) [14, 15].
Importantly, protein aggregation can result from either (1) a rare mutation in the disease-related gene encoding the protein, or (2) posttranslational changes to the protein engendered by nitrosative/oxidative stress, which may well account for the more common sporadic cases of the disease [16]. Therefore, a key theme of this article is the hypothesis that nitrosative and oxidative stress contribute to protein misfolding in the brains of the majority of neurodegenerative patients. In this review, we discuss specific examples showing that S-nitrosylation of (1) ubiquitin E3 ligases such as parkin or (2) endoplasmic reticulum chaperones such as protein-disulfide isomerase (PDI) is critical for the accumulation of misfolded proteins in neurodegenerative diseases such as PD and other conditions [17–20]. We also discuss the neuroprotective mechanism of action of NMDA open-channel blockers like memantine and NO-related drugs for the treatment of neurodegenerative disorders [11, 21].
Protein misfolding and aggregation in neurodegenerative diseases
In general, protein aggregates do not accumulate in unstressed, healthy neurons due in part to the existence of cellular ‘quality control machineries.’ For example, molecular chaperones are believed to provide a defense mechanism against the toxicity of misfolded proteins because chaperones can prevent inappropriate interactions within and between polypeptides, and can promote refolding of proteins that have been misfolded because of cell stress. In addition to the quality control of proteins provided by molecular chaperones, the ubiquitin-proteasome system (UPS) and autophagy/lysosomal degradation are involved in the clearance of abnormal or aberrant proteins. When chaperones cannot repair misfolded proteins, they may be tagged via addition of polyubiquitin chains for degradation by the proteasome. In neurodegenerative conditions, intra- or extra-cellular protein aggregates are thought to accumulate in the brain as a result of a decrease in molecular chaperone or proteasome activities (Fig. 1). In fact, several mutations that disturb the activity of molecular chaperones or UPS-associated enzymes can cause neurodegeneration [2, 22, 23]. Along these lines, postmortem samples from the substantia nigra of PD patients (versus non-PD controls) manifest a significant reduction in proteasome activity [24]. Moreover, overexpression of the molecular chaperone HSP70 can prevent neurodegeneration in vivo in models of PD [25].
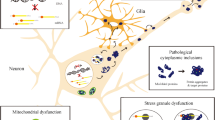
Protein misfolding and protein quality control. Misfolded proteins are thought to form small toxic oligomers. Molecular chaperones can facilitate proper protein folding and thus prevent protein aggregation. Alternatively, if a higher molecular mass complex of oligomers forms, this may offer protection by avoiding the toxicity caused by soluble oligomers. UPS-mediated proteasome degradation can serve as a rapid and efficient pathway to remove misfolded proteins. Additionally, macroautophagy can enhance the clearance of misfolded proteins that are poor substrates for the proteasome, such as highly toxic oligomers and aggregates [174]. Under pathological conditions, reduced activity of protein quality control systems, such as molecular chaperones, UPS, and autophagy, leads to accumulation of toxic oligomers, which in turn contributes to the progression of ‘protein conformational diseases’
Historically, lesions that contain aggregated proteins were considered to be pathogenic. Recently, several lines of evidence have suggested that aggregates are formed through a complex multi-step process by which misfolded proteins assemble into inclusion bodies; currently, soluble (micro-) oligomers of these aberrant proteins are thought to be the most toxic forms via interference with normal cell activities, while frank macroscopic aggregates may be an attempt by the cell to wall off potentially toxic material [12, 26]. Additionally, at least in yeast and cell culture models, highly toxic aggregates accumulate in a perivacuolar compartment where the autophagic pathway catalyzes clearance of aggresomes. Relatively less-toxic misfolded proteins are sequestered in juxtanuclear inclusions, which often contain molecular chaperones and proteasomes as part of the quality control machinery [27].
NMDA receptor-mediated glutamatergic signaling pathways induce Ca2+ influx and generation of RNS/ROS
It is well known that the amino-acid glutamate is the major excitatory neurotransmitter in the brain. Glutamate is present in high concentrations in the adult central nervous system and is released for milliseconds from nerve terminals in a Ca2+-dependent manner. After glutamate enters synaptic cleft, it diffuses across the cleft to interact with its corresponding receptors on the postsynaptic face of an adjacent neuron. Excitatory neurotransmission is necessary for the normal development and plasticity of synapses, and for some forms of learning or memory; however, excessive activation of glutamate receptors is implicated in neuronal damage in many neurological disorders ranging from acute hypoxic-ischemic brain injury to chronic neurodegenerative diseases. It is currently thought that overstimulation of extrasynaptic NMDA receptors mediate this neuronal damage, while, in contrast, synaptic activity predominantly activates survival pathways [28–30]. Intense hyperstimulation of excitatory receptors leads to necrotic cell death, but more mild or chronic overstimulation can result in apoptotic or other forms of cell death [31–33].
There are two large families of glutamate receptors in the nervous system, ionotropic receptors (representing ligand-gated ion channels) and metabotropic receptors (coupled to G-proteins). Ionotropic glutamate receptors are further divided into three broad classes, NMDA receptors, α-amino-3-hydroxy-5 methyl-4-isoxazole propionic acid (AMPA) receptors, and kainate receptors, which are each named after synthetic ligands that can selectively activate these receptors. The NMDA receptor has attracted attention for a long period of time because it has several properties that set it apart from other ionotrophic glutamate receptors. One such characteristic, in contrast to most AMPA and kainate receptors, is that NMDA receptor-coupled channels are highly permeable to Ca2+, thus permitting Ca2+ entry after ligand binding if the cell is depolarized in order to relieve block of the receptor-associated ion channel by Mg2+ [34, 35]. Subsequent binding of Ca2+ to various intracellular molecules can lead to many significant consequences. In particular, excessive activation of NMDA receptors leads to the production of damaging free radicals (e.g., NO and ROS) and other enzymatic processes, contributing to cell death [10, 15, 32, 33, 36, 37].
Excessive activation of glutamate receptors is implicated in neuronal damage in many neurological disorders. John Olney coined the term “excitotoxicity” to describe this phenomenon [38, 39]. This form of toxicity is mediated at least in part by excessive activation of NMDA-type receptors [10, 11, 40], resulting in excessive Ca2+ influx through a receptor’s associated ion channel. Increased levels of neuronal Ca2+, in conjunction with the Ca2+-binding protein CaM, trigger the activation of nNOS and subsequent generation of NO from the amino acid l-arginine [12, 41] (Fig. 2). NO is a gaseous free radical (thus highly diffusible) and a key molecule that plays a vital role in normal signal transduction but in excess can lead to neuronal cell damage and death. The discrepancy of NO effects on neuronal survival can also be caused by the formation of different NO species or intermediates: NO radical (NO·), nitrosonium cation (NO+), nitroxyl anion (NO−, with high energy singlet and lower energy triplet forms) [15]. Three subtypes of NOS have been identified; two constitutive forms of NOS–nNOS and endothelial NOS (eNOS)—take their names from the cell type in which they were first found. The name of the third subtype—inducible NOS (iNOS)—indicates that expression of the enzyme is induced by acute inflammatory stimuli. For example, activated microglia may produce neurotoxic amounts of NO via iNOS expression in various neurodegenerative diseases. All three isoforms are widely distributed in the brain. Each NOS isoform contains an oxidase domain at its amino-terminal end and a reductase domain at its carboxy-terminal end, separated by a Ca2+/CaM binding site [12, 41–44]. Constitutive and inducible NOS are also further distinguished by CaM binding: nNOS and eNOS bind CaM in a reversible Ca2+-dependent manner. In contrast, iNOS binds CaM so tightly at resting intracellular Ca2+ concentrations that its activity does not appear to be affected by transient variations in Ca2+ concentration. Interestingly, in order to terminate iNOS-mediated NO production, microglia may redistribute iNOS to the aggresome for inactivation [45].
Recent studies further pointed out the potential connection between ROS/RNS and mitochondrial dysfunction in neurodegenerative diseases, especially in PD [9, 46]. Pesticide and other environmental toxins that inhibit mitochondrial complex I result in oxidative and nitrosative stress, and consequent aberrant protein accumulation [17, 18, 20, 47, 48]. Administration to animal models of complex I inhibitors, such as MPTP, 6-hydroxydopamine, rotenone, and paraquat, which result in overproduction of ROS/RNS, reproduces many of the features of sporadic PD, such as dopaminergic neuron degeneration, up-regulation and aggregation of α-synuclein, Lewy body-like intraneuronal inclusions, and behavioral impairment [9, 46]. In addition, it has recently been proposed that mitochondrial cytochrome oxidase can produce NO in a nitrite (NO2 −)- and pH-dependent but non-Ca2+-dependent manner [49].
Increased nitrosative and oxidative stress are associated with chaperone and proteasomal dysfunction, resulting in accumulation of misfolded aggregates [16, 50]. However, until recently little was known regarding the molecular and pathogenic mechanisms underlying contribution of NO to the formation of inclusion bodies such as amyloid plaques in AD or Lewy bodies in PD.
Nitrosative stress regulates protein misfolding and neuronal cell death
Extreme nitrosative/oxidative stress can facilitate protein misfolding and aggregation—and very likely vice-versa. This relationship between ROS/RNS and protein misfolding is thought to play a role as a pathogenic trigger of neurodegenerative diseases, although the exact mechanism underlying ROS/RNS-mediated aggregate formation has remained elusive. Recent scientific advances, however, have implied that NO-related species may significantly participate in the process of protein misfolding through protein S-nitrosylation (and possibly nitration) under degenerative conditions.
Early investigations indicated that NO participates in cellular signaling pathways, which regulate broad aspects of brain function, including synaptic plasticity, normal development, and neuronal cell death [36, 51–53]. In general, NO exerts physiological and some pathophysiological effects via stimulation of guanylate cyclase to form cyclic guanosine-3′,5′-monophosphate (cGMP) or through S-nitros(yl)ation of regulatory protein thiol groups [13, 15, 50, 54–56]. S-Nitrosylation is the covalent addition of an NO group to a critical cysteine thiol/sulfhydryl (RSH or, more properly, thiolate anion, RS−) to form an S-nitrosothiol derivative (R-SNO). Such modification modulates the function of a broad spectrum of mammalian, plant, and microbial proteins. In general, a consensus motif of amino acids comprised of nucleophilic residues (generally an acid and a base) surround a critical cysteine, which increases the cysteine sulfhydryl’s susceptibility to S-nitrosylation [57, 58]. In contrast, denitrosylating enzymes and pathways, such as those mediated by thioredoxin/thioredoxin reductase, PDI, and intracellular glutathione, can decrease the lifespan of protein SNOs [59–61]. Our group first identified the physiological relevance of S-nitrosylation by showing that NO and related RNS exert paradoxical effects via redox-based mechanisms—NO is neuroprotective via S-nitrosylation of NMDA receptors (as well as other subsequently discovered targets, including caspases), and yet can also be neurodestructive by formation of peroxynitrite (or, as later discovered, reaction with additional molecules such as MMP-9 and GAPDH) [15, 62–69]. Over the past decade, accumulating evidence has suggested that S-nitrosylation can regulate the biological activity of a great variety of proteins, in some ways akin to phosphorylation [15, 17, 18, 20, 58, 68–76]. Chemically, NO is often a good “leaving group,” facilitating further oxidation of critical thiol to disulfide bonds among neighboring (vicinal) cysteine residues or, via reaction with ROS, to sulfenic (–SOH), sulfinic (–SO2H) or sulfonic (–SO3H) acid derivatization of the protein [18, 20, 68, 77]. Alternatively, S-nitrosylation may possibly produce a nitroxyl disulfide, in which the NO group is shared by close cysteine thiols [78].
Analyses of mice deficient in either nNOS or iNOS confirmed that NO is an important mediator of cell injury and death after excitotoxic stimulation; NO generated from nNOS or iNOS is detrimental to neuronal survival [79, 80]. In addition, inhibition of NOS activity ameliorates the progression of disease pathology in animal models of PD, AD, and ALS, suggesting that excess generation of NO plays a pivotal role in the pathogenesis of several neurodegenerative diseases [81–84]. Intriguingly, levels of glutathione diminish by ~30% in the aged brain [85], potentially assisting the accumulation of SNOs in elderly. Although the involvement of NO in neurodegeneration has been widely accepted, the chemical relationship between nitrosative stress and accumulation of misfolded proteins has remained obscure. Recent findings, however, have shed light on molecular events underlying this relationship. Specifically, we recently mounted physiological and chemical evidence that S-nitrosylation modulates the (1) ubiquitin E3 ligase activity of parkin [17–19], and (2) chaperone and isomerase activities of PDI [20], contributing to protein misfolding and neurotoxicity in models of neurodegenerative disorders.
Additionally, peroxynitrite-mediated nitration of tyrosine residue(s) may potentially contribute to dysfunctional protein folding and neuronal cell injury. For instance, nitration of α-synuclein and tau effects oligomer formation in vitro. Furthermore, it has been reported that nitrated α-synuclein and tau selectively accumulate in inclusion bodies in PD and neurofibrillary tangles in AD brains [86–89]. Collectively, these findings support the proposition that S-nitrosylation and possibly nitration can influence aggregate formation and neurotoxicity.
Parkin and the UPS
Recent studies on rare genetic forms of PD have found that mutations in the genes encoding parkin (PARK2), PINK1 (PARK6), α-synuclein (PARK1/4), DJ-1 (PARK7), ubiquitin C-terminal hydrolase L1 (UCH-L1) (PARK5), leucine-rich repeat kinase-2 (LRRK2) (PARK8), or ATP13A2 (PARK9) are associated with PD pathology [90–97]. The discovery that mutations in these genes predispose patients to very rare familial forms of PD have allowed us to begin to understand the mechanism of protein aggregation and neuronal loss in the more common sporadic forms of PD. For instance, the identification of α-synuclein as a familial PD gene led to the recognition that one of the major constituents of Lewy bodies in sporadic PD brains is α-synuclein. In addition, identification of errors in the genes encoding parkin (a ubiquitin E3 ligase) and UCH-L1 in rare familial forms of PD has implicated possible dysfunction of the UPS in the pathogenesis of sporadic PD as well. The UPS represents an important mechanism for proteolysis in mammalian cells. Formation of polyubiquitin chains constitutes the signal for proteasomal attack and degradation. An isopeptide bond covalently attaches the C terminus of the first ubiquitin in a polyubiqutin chain to a lysine residue in the target protein. The cascade of activating (E1), conjugating (E2), and ubiquitin-ligating (E3) type enzymes catalyzes the conjugation of the ubiquitin chain to proteins. In addition, individual E3 ubiquitin ligases play a key role in the recognition of specific substrates [98].
Mutations in the parkin gene can cause autosomal recessive juvenile Parkinsonism (ARJP), accounting for some cases of hereditary PD manifest in young patients with onset beginning anywhere from the teenage years through the 40s [22, 90, 99]. Parkin is a member of a large family of E3 ubiquitin ligases that are related to one another by the presence of RING finger domains. Parkin contains a total of 35 cysteine residues, the majority of which reside within its RING domains, which coordinate a structurally important zinc atom often involved in catalysis [100]. Parkin has two RING finger domains separated by an “in between RING” (IBR) domain. This motif allows parkin to recruit substrate proteins as well as an E2 enzyme (e.g., UbcH7, UbcH8, or UbcH13). Point mutations, stop mutations, truncations, and deletions in both alleles of the parkin gene will eventually cause dysfunction in its activity and are responsible for many cases of ARJP as well as rare adult forms of PD. Parkin mutations usually do not facilitate the formation of Lewy bodies, although there is at least one exception—familial PD patients with the R275W parkin mutant manifest Lewy bodies [101]. Biochemical characterization of parkin mutants show that not all parkin mutations result in loss of parkin E3 ligase activity; some of the familial-associated parkin mutants (e.g., the R275W mutant) have increased ubiquitination activity compared to wild-type [102–104]. Additionally, parkin can mediate the formation of non-classical and “non-degradative” lysine 63-linked polyubiquitin chains [105, 106]. Likewise, parkin can mono-ubiquitinates Eps15, HSP70, and itself possibly at the multiple sites. This finding may explain how some parkin mutations induce formation of Lewy bodies and why proteins are stabilized within the inclusions.
Possible mechanism whereby S-nitrosylated species contribute to the accumulation of aberrant proteins and neuronal damage. NMDAR hyperactivation triggers generation of NO/ROS and cytochrome C release from mitochondria associated with subsequent activation of caspases, causing neuronal cell damage and death. S-Nitrosylation of parkin (forming SNO-PARK) and PDI (forming SNO-PDI) can contribute to neuronal cell injury in part by triggering accumulation of misfolded proteins. Memantine and NitroMemantine preferentially block excessive (pathological/extrasynatpic) NMDAR activity while relatively sparing normal (physiological/synaptic) activity
Several putative target substrates have been identified for parkin E3 ligase activity. One group has reported that mutant parkin failed to bind glycosylated α-synuclein for ubiquitination, leading to α-synuclein accumulation [107], but most authorities do not feel that α-synuclein is a direct substrate of parkin. Synphilin-1 (α-synuclein interacting protein), on the other hand, is considered to be a substrate for parkin ubiquitination, and it is included in Lewy body-like inclusions in cultured cells when co-expressed with α-synuclein [108]. Other substrates for parkin include parkin-associated endothelin receptor-like receptor (Pael-R) [109], cell division control related protein (CDCrel-1) [110], cyclin E [111], p38 tRNA synthase [112], and synaptotagmin XI [113], α/β tubulin heterodimers [114], as well as possibly parkin itself (auto-ubiquitination). It is generally accepted that accumulation of these substrates can lead to disastrous consequences for the survival of dopaminergic neurons in familial PD and possibly also in sporadic PD. Therefore, characterization of potential regulators that affect parkin E3 ligase activity may reveal important molecular mechanisms for the pathogenesis of PD. Heretofore, two cellular components have been shown to regulate the substrate specificity and ubiquitin E3 ligase activity of parkin. The first represents posttranslational modification of parkin through S-nitrosylation or phosphorylation [115], and the second, binding partners of parkin, such as CHIP [116] and BAG5 [117]. CHIP enhances the ability of parkin to inhibit cell death through up-regulation of parkin-mediated ubiquitination, while BAG5-mediated inhibition of parkin E3 ligase activity facilitates neuronal cell death. In addition, several groups have recently reported that parkin-mediated mono-ubiquitination could contribute to neuronal survival via a proteasome-independent pathway [103, 104, 118, 119]. For example, parkin mono-ubiquitinates the epidermal growth factor receptor (EGFR)-associated protein, Eps15, leading to inhibition of EGFR endocytosis [118]. The resulting prolongation of EGFR signaling via the phosphoinositide-3 kinase/Akt (PKB) signaling pathway is postulated to enhance neuronal survival.
Another important molecule that links aberrant UPS activity and PD is the ubiquitin hydrolase Uch-L1, a deubiquitinating enzyme that recycles ubiquitin. Autosomal dominant mutations of Uch-L1 have been identified in two siblings with PD [94]. Interestingly, a recent study suggested that a novel ubiquitin-ubiquitin ligase activity of Uch-L1 might also be important in the pathogenesis of PD [120]. Additional mutations in α-synuclein, DJ-1, PINK1, and LRRK2 may contribute to UPS dysfunction and subsequently lead to PD.
S-Nitrosylation and parkin
PD is the second most prevalent neurodegenerative disease and is characterized by the progressive loss of dopamine neurons in the substantia nigra pars compacta. Appearance of Lewy bodies that contain misfolded and ubiquitinated proteins generally accompanies the loss of dopaminergic neurons in the PD brain. Such ubiquitinated inclusion bodies are the hallmark of many neurodegenerative disorders. Age-associated defects in intracellular proteolysis of misfolded or aberrant proteins might lead to accumulation and ultimately deposition of aggregates within neurons or glial cells. Although such aberrant protein accumulation had been observed in patients with genetically encoded mutant proteins, recent evidence from our laboratory suggests that nitrosative and oxidative stress are potential causal factors for protein accumulation in the much more common sporadic form of PD. As illustrated below, nitrosative/oxidative stress, commonly found during normal aging, can mimic rare genetic causes of disorders, such as PD, by promoting protein misfolding in the absence of a genetic mutation [17–19]. For example, S-nitrosylation and further oxidation of parkin or Uch-L1 result in dysfunction of these enzymes and thus of the UPS [17, 18, 121–124]. We and others recently discovered that nitrosative stress triggers S-nitrosylation of parkin (forming SNO-parkin) not only in rodent models of PD but also in the brains of human patients with PD and the related α-synucleinopathy, DLBD (diffuse Lewy body disease). SNO-parkin initially stimulates ubiquitin E3 ligase activity, resulting in enhanced ubiquitination as observed in Lewy bodies, followed by a decrease in enzyme activity, producing a futile cycle of dysfunctional UPS [18, 19, 105] (Fig. 2). We also found that rotenone led to the generation of SNO-parkin and thus dysfunctional ubiquitin E3 ligase activity. Moreover, S-nitrosylation appears to compromise the neuroprotective effect of parkin [17]. These mechanisms involve S-nitrosylation of critical cysteine residues in the first RING domain of parkin [18]. Nitrosative and oxidative stress can also alter the solubility of parkin via posttranslational modification of cysteine residues, which may concomitantly compromise its protective function [125–127]. Additionally, it is likely that other ubiquitin E3 ligases with RING-finger thiol motifs are S-nitrosylated in a similar manner to parkin to affect their enzymatic function; hence, S-nitrosylation of E3 ligases may be involved in a number of degenerative conditions.
The neurotransmitter dopamine (DA) may also impair parkin activity and contribute to neuronal demise via the modification of cysteine residue(s) [128]. DA can be oxidized to DA quinone, which can react with and inactivate proteins through covalent modification of cysteine sulfhydryl groups; peroxynitrite has been reported to promote oxidation of DA to form dopamine quinone [129]. DA quinone can preferentially attack cysteine residues (C268 and C323) in the RING1 and IBR domains of parkin, forming a covalent adduct that abrogates its E3 ubiquitin ligase activity [126, 128]. DA quinone also reduces the solubility of parkin, possibly inducing parkin misfolding after disruption of the RING-IBR-RING motif. Therefore, oxidative/nitrosative species may either directly or indirectly contribute to altered parkin activity within the brain, and subsequent loss of parkin-dependent neuroprotection results in increased cell death.
The unfolded protein response (UPR) and PDI
The ER normally participates in protein processing and folding but undergoes a stress response when immature or misfolded proteins accumulate [130–133]. ER stress stimulates two critical intracellular responses. The first represents expression of chaperones that prevent protein aggregation via the UPR, and is implicated in protein refolding, post-translational assembly of protein complexes, and protein degradation. This response is believed to contribute to adaptation during altered environmental conditions, promoting maintenance of cellular homeostasis. At least three ER transmembrane sensor proteins are involved in the UPR: PKR-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1). The activation of all three proximal sensors results in the attenuation of protein synthesis via eukaryotic initiation factor-2 (eIF2) kinase and increased protein folding capacity of the ER [134–137]. The second ER stress response, termed ER-associated degradation (ERAD), specifically recognizes terminally misfolded proteins for retro-translocation across the ER membrane to the cytosol, where they can be degraded by the UPS. Additionally, although severe ER stress can induce apoptosis, the ER withstands relatively mild insults via expression of stress proteins such as glucose-regulated protein (GRP) and PDI. These proteins behave as molecular chaperones that assist in the maturation, transport, and folding of secretory proteins.
During protein folding in the ER, PDI can introduce disulfide bonds into proteins (oxidation), break disulfide bonds (reduction), and catalyze thiol/disulfide exchange (isomerization), thus facilitating disulfide bond formation, rearrangement reactions, and structural stability [138]. PDI has four domains that are homologous to thioredoxin (TRX) (termed a, b, b′, and a′). Only two of the four TRX-like domains (a and a′) contain a characteristic redox-active CXXC motif, and these two-thiol/disulfide centers function as independent active sites [139–142]. These active-site cysteines can be found in two different redox states: oxidized (disulfide) or reduced (free sulfhydryls or thiols). During oxidation of a target protein, oxidized PDI catalyzes disulfide formation in the substrate protein, resulting in the reduction of PDI. In contrast, the reduced form of the active-site cysteines can initiate isomerization by attacking the disulfide of a substrate protein and forming a transient intermolecular disulfide bond. As a consequence, an intramolecular disulfide rearrangement occurs within the substrate itself, resulting in the generation of reduced PDI. The recently determined structure of yeast PDI revealed that the four TRX-like domains form a twisted “U” shape with the two active sites facing each other on opposite sides of the “U” [143]. Hydrophobic residues line the inside surface of the “U,” facilitating interactions between PDI and misfolded proteins. Specifically, the b′ domain of PDI constitutes a part of the base of the “U” shaped structure and contributes to the efficient binding of misfolded proteins [144]. Several mammalian PDI homologues, such as ERp57 and PDIp, also localize to the ER and may manifest similar functions [145, 146]. Increased expression of PDIp in neuronal cells under conditions mimicking PD suggest the possible contribution of PDIp to neuronal survival [145]. Additionally, ERdj5, an ER reductase that contains four TRX-like domains, forms a functional ERAD complex with GRP, promoting the degradation of misfolded proteins via ERAD [147].
In many neurodegenerative disorders and cerebral ischemia, the accumulation of immature and denatured proteins results in ER dysfunction [145, 148–150], but up-regulation of PDI represents an adaptive response promoting protein refolding and may offer neuronal cell protection [145, 146, 151, 152]. In a recent study, we reported that the S-nitrosylation of PDI (to form SNO-PDI) disrupts its neuroprotective role [20].
S-Nitrosylation of PDI mediates protein misfolding and neurotoxicity in cell models of PD or AD
Disturbance of Ca2+ homeostasis within the ER plays a critical role in the accumulation of misfolded proteins and ER stress because the function of several ER chaperones requires high concentrations of Ca2+. In addition, it is generally accepted that excessive generation of NO can contribute to activation of the ER stress pathway, at least in some cell types [153, 154]. Molecular mechanisms by which NO induces protein misfolding and ER stress, however, have remained enigmatic until recently. The ER normally manifests a relatively positive redox potential in contrast to the highly reducing environment of the cytosol and mitochondria. This redox environment can influence the stability of protein S-nitrosylation and oxidation reactions [155]. S-Nitrosylation can enhance the activity of the ER Ca2+ channel-ryanodine receptor [156], which may provide a clue to how NO disrupts Ca2+ homeostasis in the ER and activates the cell death pathway. Interestingly, we have recently reported that excessive NO can also lead to S-nitrosylation of the active-site thiol groups of PDI, and this reaction inhibits both its isomerase and chaperone activities [20]. Mitochondrial complex I insult by rotenone can also result in S-nitrosylation of PDI in cell culture models. Moreover, we found that PDI is S-nitrosylated in the brains of virtually all cases examined of sporadic AD and PD. Under pathological conditions, it is possible that both cysteine sulfhydryl groups in the TRX-like domains of PDI form SNOs. Unlike formation of a single SNOs which is commonly seen after de-nitrosylation reactions catalyzed by PDI [72], dual nitrosylation may be relatively more stable and prevent subsequent disulfide formation on PDI. Therefore, we speculate that these pathological S-nitrosylation reactions on PDI are more easily detected during neurodegenerative conditions. Additionally, it is possible that vicinal (nearby) cysteine thiols reacting with NO can form nitroxyl disulfide [78], and such reaction may potentially occur in the catalytic side of PDI to inhibit enzymatic activity. In order to determine the consequences of S-nitrosylated PDI (SNO-PDI) formation in neurons, we exposed cultured cerebrocortical neurons to neurotoxic concentrations of NMDA, thus inducing excessive Ca2+ influx and consequent NO production from nNOS. Under these conditions, we found that PDI was S-nitrosylated in a NOS-dependent manner. SNO-PDI formation led to the accumulation of polyubiquitinated/misfolded proteins and activation of the UPR. Moreover, S-nitrosylation abrogated the inhibitory effect of PDI on aggregation of proteins observed in Lewy body inclusions [20, 108]. S-Nitrosylation of PDI also prevented its attenuation of neuronal cell death triggered by ER stress, misfolded proteins, or proteasome inhibition (Fig. 2). Further evidence suggested that SNO-PDI may in effect transport NO to the extracellular space, where it could conceivably exert additional adverse effects [72]. Additionally, NO can possibly mediate cell death or injury via S-nitrosylation or nitration reactions on other TRX-like proteins, such as TRX itself and glutaredoxin [71, 157, 158].
In addition to PDI, S-nitrosylation is likely to affect critical thiol groups on other chaperones, such as HSP90 in the cytoplasm [159] and possibly GRP in the ER. Normally, HSP90 stabilizes misfolded proteins and modulates the activity of cell signaling proteins including NOS and calcineurin [2]. In AD brains, levels of HSP90 are increased in both the cytosolic and membranous fractions, where HSP90 is thought to maintain tau and Aβ in a soluble conformation, thereby averting their aggregation [160, 161]. Martínez-Ruiz et al. [159] recently demonstrated that S-nitrosylation of HSP90 can occur in endothelial cells, and this modification abolishes its ATPase activity, which is required for its function as a molecular chaperone. These studies imply that S-nitrosylation of HSP90 in neurons of AD brains may contribute to the accumulation of tau and Aβ aggregates.
The UPS is apparently impaired in the aging brain. Additionally, inclusion bodies similar to those found in neurodegenerative disorders can appear in brains of normal aged individuals or those with subclinical manifestations of disease [162]. These findings suggest that the activity of the UPS and molecular chaperones may decline in an age-dependent manner [163]. Given that we have not found detectable quantities of SNO-parkin and SNO-PDI in normal aged brain [17, 18, 20] we speculate that S-nitrosylation of these and similar proteins may represent a key event that contributes to susceptibility of the aging brain to neurodegenerative conditions.
Potential treatment of excessive NMDA-induced Ca2+ influx and free radical generation
One mechanism that could potentially curtail excessive Ca2+ influx and resultant overstimulation of nNOS activity would be inhibition of NMDA receptors. Until recently, however, drugs in this class blocked virtually all NMDA receptor activity, including physiological activity, and therefore manifest unacceptable side effects by inhibiting normal functions of the receptor. For this reason, many previous NMDA receptor antagonists have disappointingly failed in advanced clinical trials conducted for a number of neurodegenerative disorders. In contrast, studies in our laboratory first showed that the adamantine derivative, memantine, preferentially blocks excessive (pathological) NMDA receptor activity while relatively sparing normal (physiological) activity (Fig. 2). Memantine does this in a surprising fashion because of its low (micromolar) affinity, even though its actions are quite selective for the NMDA receptor at that concentration. “Apparent” affinity of a drug is determined by the ratio of its “on-rate” to its “off-rate” for the target. The on-rate is not only a property of drug diffusion and interaction with the target, but also the drug’s concentration. In contrast, the off-rate is an intrinsic property of the drug-receptor complex, unaffected by drug concentration. A relatively fast off-rate is a major contributor to memantine’s low affinity for the NMDA receptor. The inhibitory activity of memantine involves blockade of the NMDA receptor-associated ion channel when it is excessively open (termed open-channel block). The unique and subtle difference of the memantine blocking sites in the channel pore may explain the advantageous properties of memantine action.
Also critical for the clinical tolerability of memantine is its uncompetitive mechanism of action. An uncompetitive antagonist can be distinguished from a noncompetitive antagonist, which acts allosterically at a noncompetitive site, i.e., at a site other than the agonist-binding site. An uncompetitive antagonist is defined as an inhibitor whose action is contingent upon prior activation of the receptor by the agonist. Hence, the same amount of antagonist blocks higher concentrations of agonist relatively better than lower concentrations of agonist. Some open-channel blockers function as pure uncompetitive antagonists, depending on their exact properties of interaction with the ion channel. This uncompetitive mechanism of action coupled with a relatively fast off-rate from the channel yields a drug that preferentially blocks NMDA receptor-operated channels when they are excessively open while relatively sparing normal neurotransmission. In fact, the relatively fast off-rate is a major contributor to a drug like memantine’s low affinity for the channel pore. While many factors determine a drug’s clinical efficacy and tolerability, it appears that the relatively rapid off rate is a predominant factor in memantine’s tolerability in contrast to other NMDA-type receptor antagonists.[11, 40] Thus, the critical features of memantine’s mode of action are its uncompetitive mechanism and fast off-rate, or what we call a UFO drug—a drug that is present at its site of inhibitory action only when you need it and then quickly disappears.
Interestingly, memantine, which is chemically an adamantine, was first synthesized and patented in 1968 by Eli Lilly and Company, as described in the Merck Index. Memantine has been used for many years in Europe to treat PD, spasticity, convulsions, vascular dementia, and later AD [164, 165]. Interestingly, the efficacy of adamantine-type drugs in the brain was first discovered by serendipity in a patient taking amantadine for influenza (amantadine is chemically similar to memantine but lacks two side groups chains composed of methyl groups). This led scientists to believe that these drugs were dopaminergic or possibly anti-cholinergic, although, as stated above, we later discovered that memantine acts as an open-channel blocker of NMDA receptor (NMDAR)-coupled channel pore; memantine is more potent in this action than amantadine. A large number of studies with in vitro and in vivo animal models demonstrated that memantine protects cerebrocortical neurons, cerebellar neurons, and retinal neurons from NMDAR-mediated excitotoxic damage [166–170]. Importantly, in a rat stroke model, memantine, which was given up to 2 h after the ischemic event, reduced the amount of brain damage by ~50% [167, 168]. Furthermore, regulatory agencies in both Europe and the USA recently voted its approval as the first treatment for moderate-to-severe AD. It is currently under study for a number of other neurodegenerative disorders, including HIV-associated dementia, Huntington’s disease, ALS and also depression.
As promising as the results with memantine are, we are continuing to pursue ways to use additional modulatory sites on the NMDA receptor to block excitotoxicity even more effectively and safely than memantine alone. New approaches in this regard are explored below.
Future therapeutics: NitroMemantines
NitroMemantines are second-generation memantine derivatives that are designed to have enhanced neuroprotective efficacy without sacrificing clinical tolerability. S-Nitrosylation site(s) is located on the extracellular domain of the NMDA receptor, and S-nitrosylation of this site, i.e., NO reaction with the sulfhydryl group of a critical cysteine residue, down-regulates (but does not completely shut off) receptor activity [11, 21]. The drug nitroglycerin, which generates NO-related species, can act at this site to limit excessive NMDA receptor activity. In fact, in rodent models, nitroglycerin can limit ischemic damage [171], and there is some evidence that patients taking nitroglycerin for other medical reasons may be resistant to glaucomatous visual field loss [172]. Consequently, we carefully characterized the S-nitrosylation sites on the NMDA receptor in order to determine if we could design a nitroglycerin-like drug that could be more specifically targeted to the receptor. In brief, we found that five different cysteine residues on the NMDA receptor could interact with NO. One of these, located at cysteine residue #399 (Cys399) on the NR2A subunit of the NMDA receptor, mediates ≥90% of the effect of NO under our experimental conditions [67]. From crystal structure models and electrophysiological experiments, we further found that NO binding to the NMDA receptor at Cys399 may induce a conformational change in the receptor protein that makes glutamate and Zn2+ bind more tightly to the receptor. The enhanced binding of glutamate and Zn2+ in turn causes the receptor to desensitize and, consequently, the ion channel to close [76]. Electrophysiological studies have demonstrated this inhibitory effect of NO on the NMDA receptor-associated channel [15, 55, 67]. Moreover, as the oxygen tension is lowered (a pO2 of 10–20 torr is found in normal brain, and even lower levels under hypoxic/ischemic conditions), the NMDA receptor becomes more sensitive to inhibition by S-nitrosylation [173].
Unfortunately, nitroglycerin itself is not very attractive as a neuroprotective agent. The same cardiovascular vasodilator effect that makes it useful in the treatment of angina could cause dangerously large drops in blood pressure in patients with dementia, stroke, traumatic injury, or glaucoma. However, the open-channel block mechanism of memantine not only leads to a higher degree of channel blockade in the presence of excessive levels of glutamate but also can be used as a homing signal for targeting drugs, e.g., the NO group, to hyperactivated, open NMDA-gated channels. We have therefore been developing combinatorial drugs (NitroMemantines) that theoretically should be able to use memantine to target NO to the nitrosylation sites of the NMDAR in order to avoid the systemic side effects of NO. Two sites of modulation would be analogous to having two volume controls on your television set for fine-tuning the audio signal.
Preliminary studies have shown NitroMemantines to be highly neuroprotective in both in vitro and in vivo animal models [11]. In fact, they appear to be more effective than memantine at lower dosage. Moreover, because of the targeting effect of the memantine moiety, NitroMemantines appear to lack the blood pressure lowering effects typical of nitroglycerin. More research still needs to be performed on NitroMemantine drugs, but by combining two clinically tolerated drugs (memantine and nitroglycerin), we have created a new, improved class of UFO drugs that should be both clinically tolerated and neuroprotective.
Conclusions
Excessive nitrosative and oxidative stress triggered by excessive NMDA receptor activation and/or mitochondrial dysfunction may result in malfunction of the UPS or molecular chaperones, thus contributing to abnormal protein accumulation and neuronal damage in sporadic forms of neurodegenerative diseases. Our elucidation of an NO-mediated pathway to dysfunction of parkin and PDI by S-nitrosylation provides a mechanistic link between free radical production, abnormal protein accumulation, and neuronal cell injury in neurodegenerative disorders such as PD. Elucidation of this new pathway may lead to the development of additional new therapeutic approaches to prevent aberrant protein misfolding by targeted disruption or prevention of nitrosylation of specific proteins such as parkin and PDI. This article also describes the action of memantine via uncompetitive antagonism of the NMDA receptor with a fast off-rate. NitroMemantines enhance the neuroprotective efficacy over memantine at a given dose owing to its additional ability to S-nitrosylate the NMDA receptor. These drugs preferentially inhibit pathologically activated NMDA receptor while preserving its normal synaptic function; thus, they are clinically tolerated. In this chapter we propose that the next generation of CNS drugs will interact with their target only during states of pathological activation and not interfere with the target if it is functioning properly. In the future, such perspectives should lead to additional novel, clinically tolerated neuroprotective therapeutics.
References
Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292:1552–1555. doi:10.1126/science.292.5521.1552
Muchowski PJ, Wacker JL (2005) Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6:11–22. doi:10.1038/nrn1587
Ciechanover A, Brundin P (2003) The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40:427–446. doi:10.1016/S0896-6273(03)00606-8
Kopito RR, Ron D (2000) Conformational disease. Nat Cell Biol 2:E207–E209. doi:10.1038/35041139
Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795. doi:10.1038/nature05292
Barnham KJ, Masters CL, Bush AI (2004) Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov 3:205–214. doi:10.1038/nrd1330
Muchowski PJ (2002) Protein misfolding, amyloid formation, and neurodegeneration: a critical role for molecular chaperones? Neuron 35:9–12. doi:10.1016/S0896-6273(02)00761-4
Emerit J, Edeas M, Bricaire F (2004) Neurodegenerative diseases and oxidative stress. Biomed Pharmacother 58:39–46. doi:10.1016/j.biopha.2003.11.004
Beal MF (2001) Experimental models of Parkinson’s disease. Nat Rev Neurosci 2:325–334. doi:10.1038/35072550
Lipton SA, Rosenberg PA (1994) Excitatory amino acids as a final common pathway for neurologic disorders. N Engl J Med 330:613–622. doi:10.1056/NEJM199403033300907
Lipton SA (2006) Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov 5:160–170. doi:10.1038/nrd1958
Bredt DS, Hwang PM, Glatt CE et al (1991) Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature 351:714–718. doi:10.1038/351714a0
Garthwaite J, Charles SL, Chess-Williams R (1988) Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature 336:385–388. doi:10.1038/336385a0
Beckman JS, Beckman TW, Chen J et al (1990) Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA 87:1620–1624. doi:10.1073/pnas.87.4.1620
Lipton SA, Choi YB, Pan ZH et al (1993) A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature 364:626–632. doi:10.1038/364626a0
Zhang K, Kaufman RJ (2006) The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology 66:S102–S109. doi:10.1212/01.wnl.0000192306.98198.ec
Chung KK, Thomas B, Li X et al (2004) S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science 304:1328–1331. doi:10.1126/science.1093891
Yao D, Gu Z, Nakamura T et al (2004) Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA 101:10810–10814. doi:10.1073/pnas.0404161101
Lipton SA, Nakamura T, Yao D et al (2005) Comment on “S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function”. Science 308:1870. doi:10.1126/science.1110135
Uehara T, Nakamura T, Yao D et al (2006) S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441:513–517. doi:10.1038/nature04782
Lipton SA (2007) Pathologically activated therapeutics for neuroprotection. Nat Rev Neurosci 8:803–808. doi:10.1038/nrn2229
Cookson MR (2005) The biochemistry of Parkinson’s disease. Annu Rev Biochem 74:29–52. doi:10.1146/annurev.biochem.74.082803.133400
Zhao L, Longo-Guess C, Harris BS et al (2005) Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet 37:974–979. doi:10.1038/ng1620
McNaught KS, Perl DP, Brownell AL, Olanow CW (2004) Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson’s disease. Ann Neurol 56:149–162. doi:10.1002/ana.20186
Auluck PK, Chan HY, Trojanowski JQ et al (2002) Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295:865–868. doi:10.1126/science.1067389
Arrasate M, Mitra S, Schweitzer ES et al (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431:805–810. doi:10.1038/nature02998
Kaganovich D, Kopito R, Frydman J (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454:1088–1095. doi:10.1038/nature07195
Hardingham GE, Fukunaga Y, Bading H (2002) Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci 5:405–414
Papadia S, Stevenson P, Hardingham NR et al (2005) Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J Neurosci 25:4279–4287. doi:10.1523/JNEUROSCI.5019-04.2005
Papadia S, Soriano FX, Leveille F et al (2008) Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat Neurosci 11:476–487. doi:10.1038/nn2071
Ankarcrona M, Dypbukt JM, Bonfoco E et al (1995) Glutamate-induced neuronal death: a succession of necrosis or apoptosis depending on mitochondrial function. Neuron 15:961–973. doi:10.1016/0896-6273(95)90186-8
Bonfoco E, Krainc D, Ankarcrona M et al (1995) Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-D-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc Natl Acad Sci USA 92:7162–7166. doi:10.1073/pnas.92.16.7162
Budd SL, Tenneti L, Lishnak T, Lipton SA (2000) Mitochondrial and extramitochondrial apoptotic signaling pathways in cerebrocortical neurons. Proc Natl Acad Sci USA 97:6161–6166. doi:10.1073/pnas.100121097
Nowak L, Bregestovski P, Ascher P et al (1984) Magnesium gates glutamate-activated channels in mouse central neurones. Nature 307:462–465. doi:10.1038/307462a0
Mayer ML, Westbrook GL, Guthrie PB (1984) Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309:261–263. doi:10.1038/309261a0
Dawson VL, Dawson TM, London ED et al (1991) Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc Natl Acad Sci USA 88:6368–6371. doi:10.1073/pnas.88.14.6368
Lafon-Cazal M, Pietri S, Culcasi M, Bockaert J (1993) NMDA-dependent superoxide production and neurotoxicity. Nature 364:535–537. doi:10.1038/364535a0
Olney JW (1969) Brain lesions, obesity, and other disturbances in mice treated with monosodium glutamate. Science 164:719–721. doi:10.1126/science.164.3880.719
Olney JW, Wozniak DF, Farber NB (1997) Excitotoxic neurodegeneration in Alzheimer disease. New hypothesis and new therapeutic strategies. Arch Neurol 54:1234–1240
Chen HS, Lipton SA (2006) The chemical biology of clinically tolerated NMDA receptor antagonists. J Neurochem 97:1611–1626. doi:10.1111/j.1471-4159.2006.03991.x
Abu-Soud HM, Stuehr DJ (1993) Nitric oxide synthases reveal a role for calmodulin in controlling electron transfer. Proc Natl Acad Sci USA 90:10769–10772. doi:10.1073/pnas.90.22.10769
Forstermann U, Boissel JP, Kleinert H (1998) Expressional control of the ‘constitutive’ isoforms of nitric oxide synthase (NOS I and NOS III). FASEB J 12:773–790
Boucher JL, Moali C, Tenu JP (1999) Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for L-arginine utilization. Cell Mol Life Sci 55:1015–1028. doi:10.1007/s000180050352
Groves JT, Wang CC (2000) Nitric oxide synthase: models and mechanisms. Curr Opin Chem Biol 4:687–695. doi:10.1016/S1367-5931(00)00146-0
Kolodziejska KE, Burns AR, Moore RH et al (2005) Regulation of inducible nitric oxide synthase by aggresome formation. Proc Natl Acad Sci USA 102:4854–4859. doi:10.1073/pnas.0500485102
Betarbet R, Sherer TB, MacKenzie G et al (2000) Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci 3:1301–1306. doi:10.1038/81834
He Y, Imam SZ, Dong Z et al (2003) Role of nitric oxide in rotenone-induced nigro-striatal injury. J Neurochem 86:1338–1345. doi:10.1046/j.1471-4159.2003.01938.x
Abou-Sleiman PM, Muqit MM, Wood NW (2006) Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci 7:207–219. doi:10.1038/nrn1868
Castello PR, David PS, McClure T et al (2006) Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab 3:277–287. doi:10.1016/j.cmet.2006.02.011
Isaacs AM, Senn DB, Yuan M et al (2006) Acceleration of amyloid beta-peptide aggregation by physiological concentrations of calcium. J Biol Chem 281:27916–27923. doi:10.1074/jbc.M602061200
O’Dell TJ, Hawkins RD, Kandel ER, Arancio O (1991) Tests of the roles of two diffusible substances in long-term potentiation: evidence for nitric oxide as a possible early retrograde messenger. Proc Natl Acad Sci USA 88:11285–11289. doi:10.1073/pnas.88.24.11285
Bredt DS, Snyder SH (1994) Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem 63:175–195. doi:10.1146/annurev.bi.63.070194.001135
Schuman EM, Madison DV (1994) Locally distributed synaptic potentiation in the hippocampus. Science 263:532–536. doi:10.1126/science.8290963
Stamler JS, Simon DI, Osborne JA et al (1992) S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci USA 89:444–448. doi:10.1073/pnas.89.1.444
Lei SZ, Pan ZH, Aggarwal SK et al (1992) Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron 8:1087–1099. doi:10.1016/0896-6273(92)90130-6
Kandel ER, O’Dell TJ (1992) Are adult learning mechanisms also used for development? Science 258:243–245. doi:10.1126/science.1411522
Stamler JS, Toone EJ, Lipton SA, Sucher NJ (1997) (S)NO signals: translocation, regulation, and a consensus motif. Neuron 18:691–696. doi:10.1016/S0896-6273(00)80310-4
Hess DT, Matsumoto A, Kim SO et al (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6:150–166. doi:10.1038/nrm1569
Benhar M, Forrester MT, Hess DT, Stamler JS (2008) Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320:1050–1054. doi:10.1126/science.1158265
Nikitovic D, Holmgren A (1996) S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J Biol Chem 271:19180–19185. doi:10.1074/jbc.271.32.19180
Romero, JM, Bizzozero, OA (2008) Intracellular glutathione mediates the denitrosylation of protein nitrosothiols in the rat spinal cord. J Neurosci Res [Epub ahead of print]
Melino G, Bernassola F, Knight RA et al (1997) S-nitrosylation regulates apoptosis. Nature 388:432–433. doi:10.1038/41237
Tenneti L, D’Emilia DM, Lipton SA (1997) Suppression of neuronal apoptosis by S-nitrosylation of caspases. Neurosci Lett 236:139–142. doi:10.1016/S0304-3940(97)00780-5
Dimmeler S, Haendeler J, Nehls M, Zeiher AM (1997) Suppression of apoptosis by nitric oxide via inhibition of interleukin-1 beta-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J Exp Med 185:601–607. doi:10.1084/jem.185.4.601
Mannick JB, Hausladen A, Liu L et al (1999) Fas-induced caspase denitrosylation. Science 284:651–654. doi:10.1126/science.284.5414.651
Kim WK, Choi YB, Rayudu PV et al (1999) Attenuation of NMDA receptor activity and neurotoxicity by nitroxyl anion, NO. Neuron 24:461–469. doi:10.1016/S0896-6273(00)80859-4
Choi YB, Tenneti L, Le DA et al (2000) Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci 3:15–21. doi:10.1038/71090
Gu Z, Kaul M, Yan B et al (2002) S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science 297:1186–1190. doi:10.1126/science.1073634
Hara MR, Agrawal N, Kim SF et al (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7:665–674. doi:10.1038/ncb1268
Jaffrey SR, Erdjument-Bromage H, Ferris CD et al (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol 3:193–197. doi:10.1038/35055104
Haendeler J, Hoffmann J, Tischler V et al (2002) Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat Cell Biol 4:743–749. doi:10.1038/ncb851
Sliskovic I, Raturi A, Mutus B (2005) Characterization of the S-denitrosation activity of protein disulfide isomerase. J Biol Chem 280:8733–8741. doi:10.1074/jbc.M408080200
Stamler JS, Singel DJ, Loscalzo J (1992) Biochemistry of nitric oxide and its redox-activated forms. Science 258:1898–1902. doi:10.1126/science.1281928
Stamler JS (1994) Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell 78:931–936. doi:10.1016/0092-8674(94)90269-0
Stamler JS, Lamas S, Fang FC (2001) Nitrosylation. The prototypic redox-based signaling mechanism. Cell 106:675–683. doi:10.1016/S0092-8674(01)00495-0
Lipton SA, Choi YB, Takahashi H et al (2002) Cysteine regulation of protein function—as exemplified by NMDA-receptor modulation. Trends Neurosci 25:474–480. doi:10.1016/S0166-2236(02)02245-2
Stamler JS, Hausladen A (1998) Oxidative modifications in nitrosative stress. Nat Struct Biol 5:247–249. doi:10.1038/nsb0498-247
Houk KN, Hietbrink BN, Bartberger MD et al (2003) Nitroxyl disulfides, novel intermediates in transnitrosation reactions. J Am Chem Soc 125:6972–6976. doi:10.1021/ja029655l
Huang Z, Huang PL, Panahian N et al (1994) Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science 265:1883–1885. doi:10.1126/science.7522345
Iadecola C, Zhang F, Casey R et al (1997) Delayed reduction of ischemic brain injury and neurological deficits in mice lacking the inducible nitric oxide synthase gene. J Neurosci 17:9157–9164
Hantraye P, Brouillet E, Ferrante R et al (1996) Inhibition of neuronal nitric oxide synthase prevents MPTP-induced parkinsonism in baboons. Nat Med 2:1017–1021. doi:10.1038/nm0996-1017
Przedborski S, Jackson-Lewis V, Yokoyama R et al (1996) Role of neuronal nitric oxide in 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP)-induced dopaminergic neurotoxicity. Proc Natl Acad Sci USA 93:4565–4571. doi:10.1073/pnas.93.10.4565
Liberatore GT, Jackson-Lewis V, Vukosavic S et al (1999) Inducible nitric oxide synthase stimulates dopaminergic neurodegeneration in the MPTP model of Parkinson disease. Nat Med 5:1403–1409. doi:10.1038/70978
Chabrier PE, Demerle-Pallardy C, Auguet M (1999) Nitric oxide synthases: targets for therapeutic strategies in neurological diseases. Cell Mol Life Sci 55:1029–1035. doi:10.1007/s000180050353
Chen TS, Richie JP Jr, Lang CA (1989) The effect of aging on glutathione and cysteine levels in different regions of the mouse brain. Proc Soc Exp Biol Med 190:399–402
Giasson BI, Duda JE, Murray IV et al (2000) Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science 290:985–989. doi:10.1126/science.290.5493.985
Reynolds MR, Berry RW, Binder LI (2007) Nitration in neurodegeneration: deciphering the “Hows” “nYs. Biochemistry 46:7325–7336. doi:10.1021/bi700430y
Reynolds MR, Reyes JF, Fu Y et al (2006) Tau nitration occurs at tyrosine 29 in the fibrillar lesions of Alzheimer’s disease and other tauopathies. J Neurosci 26:10636–10645. doi:10.1523/JNEUROSCI.2143-06.2006
Uversky VN, Yamin G, Munishkina LA et al (2005) Effects of nitration on the structure and aggregation of alpha-synuclein. Brain Res Mol Brain Res 134:84–102. doi:10.1016/j.molbrainres.2004.11.014
Kitada T, Asakawa S, Hattori N et al (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392:605–608. doi:10.1038/33416
Valente EM, Abou-Sleiman PM, Caputo V et al (2004) Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304:1158–1160. doi:10.1126/science.1096284
Polymeropoulos MH, Lavedan C, Leroy E et al (1997) Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276:2045–2047. doi:10.1126/science.276.5321.2045
Bonifati V, Rizzu P, van Baren MJ et al (2003) Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299:256–259. doi:10.1126/science.1077209
Leroy E, Boyer R, Auburger G et al (1998) The ubiquitin pathway in Parkinson’s disease. Nature 395:451–452. doi:10.1038/26652
Paisan-Ruiz C, Jain S, Evans EW et al (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44:595–600. doi:10.1016/j.neuron.2004.10.023
Zimprich A, Biskup S, Leitner P et al (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44:601–607. doi:10.1016/j.neuron.2004.11.005
Ramirez A, Heimbach A, Grundemann J et al (2006) Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet 38:1184–1191. doi:10.1038/ng1884
Ross CA, Pickart CM (2004) The ubiquitin-proteasome pathway in Parkinson’s disease and other neurodegenerative diseases. Trends Cell Biol 14:703–711. doi:10.1016/j.tcb.2004.10.006
Shimura H, Hattori N, Kubo S et al (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25:302–305. doi:10.1038/77060
Marin I, Ferrus A (2002) Comparative genomics of the RBR family, including the Parkinson’s disease-related gene parkin and the genes of the ariadne subfamily. Mol Biol Evol 19:2039–2050
Farrer M, Chan P, Chen R et al (2001) Lewy bodies and parkinsonism in families with parkin mutations. Ann Neurol 50:293–300. doi:10.1002/ana.1132
Sriram SR, Li X, Ko HS et al (2005) Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum Mol Genet 14:2571–2586. doi:10.1093/hmg/ddi292
Hampe C, Ardila-Osorio H, Fournier M et al (2006) Biochemical analysis of Parkinson’s disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet 15:2059–2075. doi:10.1093/hmg/ddl131
Matsuda N, Kitami T, Suzuki T et al (2006) Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem 281:3204–3209. doi:10.1074/jbc.M510393200
Lim KL, Chew KC, Tan JM et al (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 25:2002–2009. doi:10.1523/JNEUROSCI.4474-04.2005
Lim KL, Dawson VL, Dawson TM (2006) Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol Aging 27:524–529. doi:10.1016/j.neurobiolaging.2005.07.023
Shimura H, Schlossmacher MG, Hattori N et al (2001) Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson’s disease. Science 293:263–269. doi:10.1126/science.1060627
Chung KK, Zhang Y, Lim KL et al (2001) Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med 7:1144–1150. doi:10.1038/nm1001-1144
Imai Y, Soda M, Inoue H et al (2001) An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 105:891–902. doi:10.1016/S0092-8674(01)00407-X
Zhang Y, Gao J, Chung KK et al (2000) Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA 97:13354–13359. doi:10.1073/pnas.240347797
Staropoli JF, McDermott C, Martinat C et al (2003) Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37:735–749. doi:10.1016/S0896-6273(03)00084-9
Corti O, Hampe C, Koutnikova H et al (2003) The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet 12:1427–1437. doi:10.1093/hmg/ddg159
Huynh DP, Scoles DR, Nguyen D, Pulst SM (2003) The autosomal recessive juvenile Parkinson disease gene product, parkin, interacts with and ubiquitinates synaptotagmin XI. Hum Mol Genet 12:2587–2597. doi:10.1093/hmg/ddg269
Ren Y, Zhao J, Feng J (2003) Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J Neurosci 23:3316–3324
Yamamoto A, Friedlein A, Imai Y et al (2005) Parkin phosphorylation and modulation of its E3 ubiquitin ligase activity. J Biol Chem 280:3390–3399. doi:10.1074/jbc.M407724200
Imai Y, Soda M, Hatakeyama S et al (2002) CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol Cell 10:55–67. doi:10.1016/S1097-2765(02)00583-X
Kalia SK, Lee S, Smith PD et al (2004) BAG5 inhibits parkin and enhances dopaminergic neuron degeneration. Neuron 44:931–945. doi:10.1016/j.neuron.2004.11.026
Fallon L, Belanger CM, Corera AT et al (2006) A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol 8:834–842. doi:10.1038/ncb1441
Moore DJ, West AB, Dikeman DA et al (2008) Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem 105:1806–1819. doi:10.1111/j.1471-4159.2008.05261.x
Liu Y, Fallon L, Lashuel HA et al (2002) The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell 111:209–218. doi:10.1016/S0092-8674(02)01012-7
Nishikawa K, Li H, Kawamura R et al (2003) Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase L1 variants. Biochem Biophys Res Commun 304:176–183. doi:10.1016/S0006-291X(03)00555-2
Choi J, Levey AI, Weintraub ST et al (2004) Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem 279:13256–13264. doi:10.1074/jbc.M314124200
Chung KK, Dawson TM, Dawson VL (2005) Nitric oxide, S-nitrosylation and neurodegeneration. Cell Mol Biol Noisy-le-grand 51:247–254
Gu Z, Nakamura T, Yao D et al (2005) Nitrosative and oxidative stress links dysfunctional ubiquitination to Parkinson’s disease. Cell Death Differ 12:1202–1204. doi:10.1038/sj.cdd.4401705
Wang C, Ko HS, Thomas B et al (2005) Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet 14:3885–3897. doi:10.1093/hmg/ddi413
Wong ES, Tan JM, Wang C et al (2007) Relative sensitivity of parkin and other cysteine-containing enzymes to stress-induced solubility alterations. J Biol Chem 282:12310–12318
LaVoie MJ, Cortese GP, Ostaszewski BL, Schlossmacher MG (2007) The effects of oxidative stress on parkin and other E3 ligases. J Neurochem 103:2354–2368. doi:10.1111/j.1471-4159.2007.04911.x
LaVoie MJ, Ostaszewski BL, Weihofen A et al (2005) Dopamine covalently modifies and functionally inactivates parkin. Nat Med 11:1214–1221. doi:10.1038/nm1314
LaVoie MJ, Hastings TG (1999) Peroxynitrite- and nitrite-induced oxidation of dopamine: implications for nitric oxide in dopaminergic cell loss. J Neurochem 73:2546–2554. doi:10.1046/j.1471-4159.1999.0732546.x
Andrews DW, Johnson AE (1996) The translocon: more than a hole in the ER membrane? Trends Biochem Sci 21:365–369
Sidrauski C, Chapman R, Walter P (1998) The unfolded protein response: an intracellular signalling pathway with many surprising features. Trends Cell Biol 8:245–249. doi:10.1016/S0962-8924(98)01267-7
Szegezdi E, Logue SE, Gorman AM, Samali A (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7:880–885. doi:10.1038/sj.embor.7400779
Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory pathway. Science 286:1882–1888. doi:10.1126/science.286.5446.1882
Kaufman RJ (1999) Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13:1211–1233. doi:10.1101/gad.13.10.1211
Mori K (2000) Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101:451–454. doi:10.1016/S0092-8674(00)80855-7
Patil C, Walter P (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 13:349–355. doi:10.1016/S0955-0674(00)00219-2
Yoshida H, Matsui T, Yamamoto A et al (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891. doi:10.1016/S0092-8674(01)00611-0
Lyles MM, Gilbert HF (1991) Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry 30:613–619. doi:10.1021/bi00217a004
Edman JC, Ellis L, Blacher RW et al (1985) Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature 317:267–270. doi:10.1038/317267a0
Vuori K, Pihlajaniemi T, Myllyla R, Kivirikko KI (1992) Site-directed mutagenesis of human protein disulphide isomerase: effect on the assembly, activity and endoplasmic reticulum retention of human prolyl 4-hydroxylase in Spodoptera frugiperda insect cells. EMBO J 11:4213–4217
Ellgaard L, Ruddock LW (2005) The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep 6:28–32. doi:10.1038/sj.embor.7400311
Gruber CW, Cemazar M, Heras B et al (2006) Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci 31:455–464. doi:10.1016/j.tibs.2006.06.001
Tian G, Xiang S, Noiva R et al (2006) The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 124:61–73. doi:10.1016/j.cell.2005.10.044
Klappa P, Ruddock LW, Darby NJ, Freedman RB (1998) The b’ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J 17:927–935. doi:10.1093/emboj/17.4.927
Conn KJ, Gao W, McKee A et al (2004) Identification of the protein disulfide isomerase family member PDIp in experimental Parkinson’s disease and Lewy body pathology. Brain Res 1022:164–172. doi:10.1016/j.brainres.2004.07.026
Hetz C, Russelakis-Carneiro M, Walchli S et al (2005) The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J Neurosci 25:2793–2802. doi:10.1523/JNEUROSCI.4090-04.2005
Ushioda R, Hoseki J, Araki K et al (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321:569–572. doi:10.1126/science.1159293
Hu BR, Martone ME, Jones YZ, Liu CL (2000) Protein aggregation after transient cerebral ischemia. J Neurosci 20:3191–3199
Rao RV, Bredesen DE (2004) Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol 16:653–662. doi:10.1016/j.ceb.2004.09.012
Atkin JD, Farg MA, Turner BJ et al (2006) Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem 281:30152–30165. doi:10.1074/jbc.M603393200
Tanaka S, Uehara T, Nomura Y (2000) Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J Biol Chem 275:10388–10393. doi:10.1074/jbc.275.14.10388
Ko HS, Uehara T, Nomura Y (2002) Role of ubiquitin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J Biol Chem 277:35386–35392. doi:10.1074/jbc.M203412200
Gotoh T, Oyadomari S, Mori K, Mori M (2002) Nitric oxide-induced apoptosis in RAW 264.7 macrophages is mediated by endoplasmic reticulum stress pathway involving ATF6 and CHOP. J Biol Chem 277:12343–12350. doi:10.1074/jbc.M107988200
Oyadomari S, Takeda K, Takiguchi M et al (2001) Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci USA 98:10845–10850. doi:10.1073/pnas.191207498
Forrester MT, Benhar M, Stamler JS (2006) Nitrosative stress in the ER: a new role for S-nitrosylation in neurodegenerative diseases. ACS Chem Biol 1:355–358. doi:10.1021/cb600244c
Xu L, Eu JP, Meissner G, Stamler JS (1998) Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science 279:234–237. doi:10.1126/science.279.5348.234
Tao L, Jiao X, Gao E et al (2006) Nitrative inactivation of thioredoxin-1 and its role in postischemic myocardial apoptosis. Circulation 114:1395–1402. doi:10.1161/CIRCULATIONAHA.106.625061
Aracena-Parks P, Goonasekera SA, Gilman CP et al (2006) Identification of cysteines involved in s-nitrosylation, s-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J Biol Chem 281:40354–40368. doi:10.1074/jbc.M600876200
Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C et al (2005) S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci USA 102:8525–8530. doi:10.1073/pnas.0407294102
Kakimura J, Kitamura Y, Takata K et al (2002) Microglial activation and amyloid-beta clearance induced by exogenous heat-shock proteins. FASEB J 16:601–603
Dou F, Netzer WJ, Tanemura K et al (2003) Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci USA 100:721–726. doi:10.1073/pnas.242720499
Gray DA, Tsirigotis M, Woulfe J (2003) Ubiquitin, proteasomes, and the aging brain. Sci Aging Knowl Environ 2003:RE6
Paz Gavilan M, Vela J, Castano A et al (2006) Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol Aging 27:973–982. doi:10.1016/j.neurobiolaging.2005.05.010
Ditzler K (1991) Efficacy and tolerability of memantine in patients with dementia syndrome. A double-blind, placebo controlled trial. Arzneimittelforschung 41:773–780
Fleischhacker WW, Buchgeher A, Schubert H (1986) Memantine in the treatment of senile dementia of the Alzheimer type. Prog Neuropsychopharmacol Biol Psychiatry 10:87–93. doi:10.1016/0278-5846(86)90047-3
Parsons CG, Danysz W, Quack G (1999) Memantine is a clinically well tolerated N-methyl-D-aspartate (NMDA) receptor antagonist—a review of preclinical data. Neuropharmacology 38:735–767. doi:10.1016/S0028-3908(99)00019-2
Chen HS, Pellegrini JW, Aggarwal SK et al (1992) Open-channel block of N-methyl-D-aspartate (NMDA) responses by memantine: therapeutic advantage against NMDA receptor-mediated neurotoxicity. J Neurosci 12:4427–4436
Chen HS, Wang YF, Rayudu PV et al (1998) Neuroprotective concentrations of the N-methyl-D-aspartate open-channel blocker memantine are effective without cytoplasmic vacuolation following post-ischemic administration and do not block maze learning or long-term potentiation. Neuroscience 86:1121–1132. doi:10.1016/S0306-4522(98)00163-8
Lipton SA (1992) Memantine prevents HIV coat protein-induced neuronal injury in vitro. Neurology 42:1403–1405
Osborne NN (1999) Memantine reduces alterations to the mammalian retina, in situ, induced by ischemia. Vis Neurosci 16:45–52. doi:10.1017/S0952523899161017
Lipton SA, Wang YF (1996) NO-related species can protect from focal cerebral ischemia/reperfusion. In: Krieglstein J, Oberpichler-Schwenk H (eds) Pharmacology of cerebral ischemia. Wissenschaftliche Verlagsgesellschaft, Stuttgart, pp 183–191
Zurakowski D, Vorwerk CK, Gorla M et al (1998) Nitrate therapy may retard glaucomatous optic neuropathy, perhaps through modulation of glutamate receptors. Vis Res 38:1489–1494. doi:10.1016/S0042-6989(98)00003-0
Takahashi H, Shin Y, Cho SJ et al (2007) Hypoxia enhances S-nitrosylation-mediated NMDA receptor inhibition via a thiol oxygen sensor motif. Neuron 53:53–64. doi:10.1016/j.neuron.2006.11.023
Rubinsztein DC (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443:780–786. doi:10.1038/nature05291
Acknowledgments
This work was supported in part by a JSPS Postdoctoral Fellowship for Research Abroad (to T. N.), NIH grants P01 HD29587, R01 EY05477, R01 EY09024, the American Parkinson’s Disease Association, San Diego Chapter, and an Ellison Senior Scholars Award in Aging (to S. A. L.).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Nakamura, T., Lipton, S.A. Cell death: protein misfolding and neurodegenerative diseases. Apoptosis 14, 455–468 (2009). https://doi.org/10.1007/s10495-008-0301-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-008-0301-y