Abstract
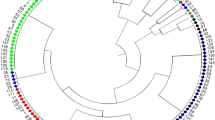
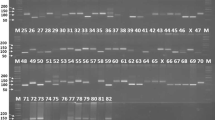
Genetic diversity in 90 Indian soybean cultivars was assessed using 45 SSR markers distributed on 20 soybean chromosomes. Forty-five SSR markers generated 232 alleles with an average of five alleles/locus. The observed frequencies of the 232 alleles ranged from 0.01 to 0.94 with an average of 0.19. The polymorphic information content (PIC) value of the SSR markers varied from 0.10 to 0.83 with an average of 0.61 and about 71% markers have a PIC value of >0.5. In this study, 54 rare alleles including 19 genotype specific alleles were also identified. The observed hetrozygosity for SSR markers ranged from 0 to 0.11 with a mean of 0.10. Cluster analysis grouped the 90 soybean cultivars into three major clusters and principal coordinates analysis (PCoA) results were similar to those of the cluster analysis. A combination of eight SSR markers successfully differentiated all 90 soybean cultivars. The population structure analysis distributed the 90 soybean genotypes into two populations with mean alpha (α) value of 0.1873. In AMOVA analysis, proportion of variation within population was high (88%), whereas only 12% occurred among populations. In cluster and structure analyses, most of the genotypes with similar pedigree were grouped together. Soybean cultivars DS228, MACS-13, LSb-1, Hardee, Improved Pelican, and Pusa-24 were the six most genetically distinct cultivars identified. The study reported a moderate genetic diversity in Indian soybean cultivars and findings would be useful to the soybean breeders in selecting genetically distinct parents for a soybean improvement program.
Similar content being viewed by others
References
Anonymous. 2014. Annual report 2013–14, AICRP on Soybean, Directorate of Soybean Research, Indore, India
Asare AT, Gowda BS, Galyuon IKA, Aboagye LL, Takrama JF, Timko MP. 2010. Assessment of the genetic diversity in cowpea (Vigna unguiculata L. Walp.) germplasm from Ghana using simple sequence repeat markers. Plant Genet. Resour. 8: 142–150
Bar-Hen A, Charcosset A, Bourgoin M, Cuiard J. 1995. Relationships between genetic markers and morphological traits in a maize inbred lines collection. Euphytica 84: 145–154
Barrett BA, Kidwell KK. 1998. AFLP-based genetic diversity assessment among wheat cultivars from the Pacific Northwest. Crop Sci. 38: 1261–1271
Bharadwaj CH, Satyavathi CT, Tiwari SP, Karmakar PG. 2002. Genetic base of soybean (Glycine max) varieties released in India as revealed by coefficient of parentage. Indian J. Agri. Sci. 72(8): 467–469
Bisen A, Khare D, Nair P, Tripathi N. 2015. SSR analysis of 38 genotypes of soybean (Glycine Max (L.)Merr.) genetic diversity in India. Physiol. Mol. Biol. Plants 21: 109–115
Botstein D, White RL, Skolnick M, Davis RW. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32: 314–331
Brown-Guedira GL, Thompson JA, Nelson RL, Warburton ML. 2000. Evaluation of genetic diversity of soybean introductions and North American ancestors using RAPD and SSR markers. Crop Sci. 40: 815–823
Burle ML, Fonseca JR, Kami JA, Gepts P. 2010. Microsatellite diversity and genetic structure among common bean (Phaseolus vulgaris L.) landraces in Brazil, a secondary center of diversity. Theor. Appl. Genet. 121: 801–813
Chauhan DK, Bhat JA, Thakur AK, Kumari S, Hussain Z, Satyawathi CT. 2015. Molecular characterization and genetic diversity assessment in soybean [Glycine max (L.) Merr.] varieties using SSR markers. Ind. J. Biotechnol. 14: 504–510.
Diwan N, Cregan PB. 1997. Automated sizing of Xuorescent-labeled simple sequence repeat (SSR) markers to assay genetic variation in soybean. Theor. Appl. Genet. 95: 723–733
Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, Dogra V, Gaikwad K, Sharma TR, Raje RS, Bandhopadhya TK, Datta S, et al. 2011. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol. 11: 17
Earl DA, von Holdt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4(2): 359–361
Fu YB, Peterson GW, Morrison MJ. 2007. Genetic diversity of Canadian soybean cultivars and exotic germplasm revealed by simple sequence repeat markers. Crop Sci. 47: 1947–1954
Gupta SK, Bansal R, Vaidya UJ, Gopalakrishna T. 2013. Assessment of genetic diversity at molecular level in mungbean (Vigna radiata (L.) Wilczek). J. Food Legumes 26(3 & 4): 19–24
Gupta SK, Gopalakrishna T. 2009. Genetic diversity analysis in blackgram (Vigna mungo (L.) Hepper) using AFLP and transferable microsatellite markers from azuki bean (Vigna angularis (Willd.) Ohwi & Ohashi). Genome 52: 120–128
Gupta SK, Gopalakrishna T. 2010. Development of unigene-derived SSR markers in cowpea (Vigna unguiculata) and their transferability to other Vigna species. Genome 53: 508–523
Halliburton R. 2004. Introduction to population genetics. Pearson/Prentice Hall, Upper Saddle River, NJ
Hymowitz T. 1969. The soybeans of the Kumaon Hills of India. Econ. Bot. 23(1): 50–54
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. 2006. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. 103: 16666–16671
Jain S, Jain R, Mccouch S. 2004. Genetic analysis of Indian aromatic and qualityrice (Oryza sativa L.) germplasm using panels of Xuorescently-labled microsatellite markers. Theor. Appl. Genet. 109: 965–977
Karp A, Edwards KJ, Bruford M, Funk S, Vosman B. 1997. Molecular technologies for biodiversity evaluation: opportunities and challenges. Nat. Biotechnol. 15: 625–628
Kumawat G, Singh G, Gireesh C, Shivakumar M, Arya M, Agarwal DK, Husain SM. 2015. Molecular characterization and genetic diversity analysis of soybean (Glycine max (L.) Merr.) germplasm accessions in India. Physiol. Mol. Biol. Plants 21(1): 101–107
Li C-D, Fatokun CA, Ubi B, Singh BB, Scoles GJ. 2001. Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci. 41: 189–197
Li Y, Guan R, Liu Z, Ma Y, Wang L, Li L, Lin F, Luan W, Chen P, Qiu L. 2008. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet. 117: 857–871
Liu K, Muse S. 2005. Powermarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129
Manjaya JG, Bapat VA. 2008. Studies on genetic divergence in soybean [Glycine max (L.) Merrill]. J. Oilseeds Res. 25: 178- 180
Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and researchan update. Bioinformatics 28: 2537–2539
Perrier X, Flori A, Bonnot F. 2003. Data analysis methods. In: P Hamon PM Seguin M, PX errier X, JC Glaszmann, Ed., Genetic diversity of cultivated tropical plants. Enfield, Science Publishers. Montpellier. pp 43–76
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A. 1996. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 2: 225–238
Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959
Russell J, Fuller J, Young G, Thomas B, Taramino G, Macaulay M, Waugh R, Powell W. 1997. Discriminating between barley genotypes using microsatellite markers. Genome 40: 442–450
Singh BB. 2006. Success of soybean in India: The Early Challenges and Pioneer Promoters. Asian Agrihist. 10: 43–53
Singh RK, Sharma RK, Singh AK, Singh VP, Singh NK, Tiwari SP, Mohapatra T. 2004. Suitability of mapped sequence tagged microsatellite site markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica 135: 135–143
Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. 2004. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109: 122–128
Tantasawat P, Trongchuen J, Prajongjai T, Jenweerawat S, Chaowiset W. 2011. SSR analysis of soybean (Glycine max (L.) Merr.) genetic relationship and variety identification in Thailand. Aust. J. Crop Sci. 5(3): 283–290
Tautz D, Renz P. 1984. Simple sequences are ubiquitous repetitive components of eukaryotic genome. Nucl. Acids Res. 12: 4127–4138
Thompson JA, Nelson RL, Vodkin LO. 1998. Identification of diverse soybean germplasm using RAPD markers. Crop Sci. 38: 1348–1355
Tiwari SP. 2014. Raising the yield ceilings in soybean - An Indian Overview. Soybean Res. 12(2): 1–43
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M. 1995. AFLP: a new technique for DNA fingerprinting. Nucl. Acids Res. 23: 4407–4414
Wang LX, Guan RX, Liu ZX, Chang RZ, Qiu LJ. 2006. Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci. 46: 1032–1038
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl. Acids Res. 18: 6531–6535
Zietkiewicz E, Rafalski A, Labuda D. 1994. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20: 176–183
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Gupta, S., Manjaya, J. Genetic diversity and population structure of Indian soybean [Glycine max (L.) Merr.] revealed by simple sequence repeat markers. J. Crop Sci. Biotechnol. 20, 221–231 (2017). https://doi.org/10.1007/s12892-017-0023-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-017-0023-0