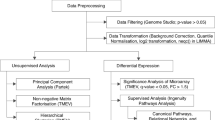
Abstract
Background and aims
HCV GT-3 has a more pronounced effect on hepatic steatosis and host lipids than other HCV genotypes and is proving less responsive to all oral interferon-free treatment with direct acting antiviral agents. As both HCV GT3 infection and NASH can result in steatosis and cirrhosis, we asked whether hepatic transcriptional profiles reflective of the host response to inflammation differed based on the etiology of injury.
Methods
Hepatic gene expression was determined for 48 pre-selected genes known to be associated with hepatic interferon signaling and lipid metabolic pathways in treatment-naïve HCV GT-3 (n = 9) and NASH (n = 14) patients.
Results
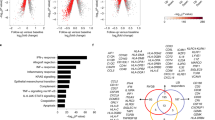
Genes with significantly higher expression in HCV included chemokines CXCL10, CXCL11 interferon IFNA2, interferon receptors IFNAR1, IL10RB negative regulators of interferon signaling SOCS3, USP18, JAK/STAT and IRF family members STAT1, STAT2, and IRF, and TGFB family members TGFB1, TGFBR1, and TGFBR2 and other ISGs like OAS2, IF127, IF144 and ISG15. HCV infection was also associated with higher expression of genes associated with lipid metabolism APOE, APOL3, SREBF1 and HMBS. Furthermore, our results suggest that, in HCV GT3-infected patients, IL28B (CC) genotype is associated with lower baseline ISG expression such as IRF9, ISG15, MX1, STAT1, CXCL10, CXCL11, and IFI27 compared to CT/TT genotype.
Conclusions
HCV GT-3 and NASH both induce hepatic steatosis and inflammation, while HCV GT-3 infection is uniquely associated with elevated transcription of hepatic ISGs and genes associated with lipid metabolism. These changes likely reflect the unique host response to HCV replication distinct from the inflammatory response induced by NASH.





Similar content being viewed by others
Abbreviations
- ALP:
-
Alkaline phosphatase
- ALT:
-
Alanine aminotransferase
- AST:
-
Serum aspartate aminotransferase
- CHOL:
-
Total cholesterol
- HCC:
-
Hepatocellular carcinoma
- HCV GT-3:
-
Hepatitis C virus GT3
- IL28 B:
-
Interleukin 28 B
- ISG:
-
Interferon stimulated genes
- NAFLD:
-
Nonalcoholic fatty liver disease
- NASH:
-
Non-alcoholic steatohepatitis
- SVR:
-
Sustained virological response
- TBILI:
-
Total bilirubin
- TRIG:
-
Triglyceride
References
MohdHanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age specific antibody to HCV seroprevalence. Hepatology 2013;57:1333–1342
Ekstedt M, Franzen LE, Mathiesen UL, Thorelius L, Holmqvist M, Bodemar G, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology 2006;44:865–873
Teli MR, James OF, Burt AD, Bennett MK, Day CP. The natural history of nonalcoholic fatty liver: a follow-up study. Hepatology 1995;22:1714–1719
Lomonaco R, Chen J, Cusi K. An endocrine perspective of nonalcoholic fatty liver disease (NAFLD). Ther Adv Endocrinol Metabol 2011;2:211–225
Lonardo A, Loria P, Adinolfi LE, Carulli N, Ruggiero G. Hepatitis C and steatosis: a reappraisal. J Viral Hepat 2006;13(2):73–80
Milić S, Lulić D, Štimac D. Non-alcoholic fatty liver disease and obesity: biochemical, metabolic and clinical presentations. World J Gastroenterol 2014;20(28):9330–9337.
Ampuero J, Romero-Gómez M, Reddy KR. Review article: HCV genotype 3—the new treatment challenge. Aliment Pharmacol Ther 2014;39(7):686–698
Jhaveri R, McHutchison J, Patel K, Qiang G, Diehl AM. Specific polymorphisms in hepatitis C virus genotype 3 core protein associated with intracellular lipid accumulation. J Infect Dis 2008;197:283–291
Rubbia-Brandt L, Quadri R, Abid K, Giostra E, Malé PJ, Mentha G, et al. Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J Hepatol 2000;33:106–115
Nkontchou G, Ziol M, Aout M, Lhabadie M, Baazia Y, Mahmoudi A et al. HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepat 2011; 18: e516–e522
Gale M Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature 2005; 436: 939–945
Negro F, Sanyal AJ. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int 2009;29(Suppl 2):26–37
Rosen HR. Emerging concepts in immunity to hepatitis C virus infection. J Clin Invest 2013;123(10):4121–4130
Lau DT, Negash A, Chen J, Crochet N, Sinha M, Zhang Y, et al. Innate immune tolerance and the role of kupffer cells in differential responses to interferon therapy among patients with HCV genotype 1 infection. Gastroenterology 2013; 144(2): 402–413
Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology 2007;46(5):1548–1563
Felmlee DJ, Hafirassou ML, Lefevre M, Baumert TF, Schuster C. Hepatitis C virus, cholesterol and lipoproteins impact for the viral life cycle and pathogenesis of liver disease. Viruses 2013;5:1292–1324
Negro F. Hepatitis C in 2013: HCV causes systemic disorders that can be cured. Nat Rev Gastroenterol Hepatol 2014;11:77–78
Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 2013;11(482–496):13
Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 2007;9:1089–1097
Katsounas A, Hubbard JJ, Wang CH, Zhang X, Dou D, Shivakumar B, et.al. High interferon-stimulated gene ISG-15 expression affects HCV treatment outcome in patients co-infected with HIV and HCV. J Med Virol 2013; 85(6): 959–993
Onomoto K, Morimoto S, Kawaguchi T, Toyoda H, Tanaka M, Kuroda M, Uno K, Kumada T, Matsuda F, Shimotohno K, Fujita T, Murakami Y. Dysregulation of IFN system can lead to poor response to pegylated interferon and ribavirin therapy in chronic hepatitis C. PLoS ONE 2011;6(5):e19799
Larrubia JR, Benito-Martínez S, Calvino M, Sanz-de-Villalobos E, Parra-Cid T. Role of chemokines and their receptors in viral persistence and liver damage during chronic hepatitis C virus infection. World J Gastroenterol 2008;14(47):7149–7159
Stättermayer AF, Scherzer T, Beinhardt S, Rutter K, Hofer H, Ferenci P. Review article: genetic factors that modify the outcome of viral hepatitis. Aliment Pharmacol Ther 2014;39(10):1059–1070
Naggie S, Osinusi A, Katsounas A, Lempicki R, Herrmann E, Thompson AJ, et al. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology 2012;56(2):444–454
Acknowledgements
We would like to thank all the patients who participated in this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding sources
National Institute of Allergy and Infectious Diseases (NIH), Bethesda, United States and Institute of Liver and biliary Sciences, New Delhi, India, supported this study.
Conflict of interest
Shikha Shrivastava, Eric G. Meissner, Emily Funk, Seerat Poonia, Virender Shokeen, Arun Thakur, Bhawna Poonia, Shiv Kumar Sarin, NirupmaTrehanpati and Shyamasundaran Kottilil declare that they have no conflicts of interest to disclose.
Ethical approval
The study protocol was approved by institutional ethical committee of Institute of Liver and Biliary Sciences (ILBS), New Delhi, and is in accordance with Declaration of Helsinki 1975.
Informed consent
All patients provided written informed consent to participate in the study.
Additional information
N. Trehanpati and S. Kottilil contributed equally to this manuscript.
Appendix
Appendix
See Table 2.
Rights and permissions
About this article
Cite this article
Shrivastava, S., Meissner, E.G., Funk, E. et al. Elevated hepatic lipid and interferon stimulated gene expression in HCV GT3 patients relative to non-alcoholic steatohepatitis. Hepatol Int 10, 937–946 (2016). https://doi.org/10.1007/s12072-016-9733-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-016-9733-6