Abstract
Glucosyltransferases (GTFs, EC.2.4.1.5) are bacterial enzymes that catalyze the polymerization of glucose residues from sucrose, leading to the production of high molecular weight glucan with α-1,3 /α-1,6 linkages. Such glucans, with many potential food and industrial applications, do not normally exist in higher plants. We fused a mutant form of the gtfD gene from Sreptococcus mutans with the maize (Zea mays L.) chloroplastic Brittle 1 transit peptide for amyloplast targeting. This construct, driven by the ubiquitin promoter, was introduced into maize by Agrobacterium-mediated transformation. We developed a novel HPLC-based method that enabled us differentially to distinguish transgene glucan from other endogenous polysaccharides in maize kernels. Using this method, we screened over 100 transgenic plants for the presence of GTF-produced glucan whose content varied between 0.8 and 14% of dry weight in the mature transgenic seeds. The mature transgenic plants were indistinguishable from wildtype plants in growth rate and morphology. Furthermore, starch granule size in the transgenic maize kernel was unaffected by the accumulation of the foreign polysaccharide. Mutation in Sh2, which encodes a subunit of ADP-glucose pyrophosphorylase, had no effect on glucan accumulation caused by gtfD expression. Our results indicated that high levels of novel carbohydrate polymer can be accumulated in crop plants through transgene technology.
Similar content being viewed by others
Introduction
Glucans are oligo- and polysaccharides composed solely of glucose molecules. They exist widely in nature and have diverse structural, physical, and chemical properties depending on the linkage between glucose units and the length of the polymer chain. There is significant diversity among α-d-glucans. Amylose and amylopectin, two components of starch granules, are both α-d-glucans but show distinct properties. Amylose is a linear molecule with α-(1→4)-linked glucose residues and amylopectin is a branched glucan containing both α-(1→4)-linkage and a low degree of α-(1→4,6)-linkages that create branch points. Starch granules with different ratios and distribution of amylose and amylopectin exhibit distinct properties regarding gelatinization temperature, retrogradation, and viscosity (Blanshard 1987) and, therefore, they have different industrial applications. Starch is the most abundant storage carbohydrate in plants and is in huge demand as a raw material for many specialty carbohydrate polymers in various food and industrial applications. However, the naturally occurring diversity in starch structures is limited which also limits starch’s potential utility. The current industrial practice of chemically modifying starch is costly and imposes undesired environmental burdens. Introduction of greater diversity in linkages within α-glucan polymers will enable the generation of specialty glucans to replace modified starches used for thickening reagents, adhesives, textile modification, and papermaking polymers with economic and environmental benefits.
Streptococcus mutans strains have been implicated as primary etiological agents in the development of human dental caries (Hanada and Kuramitsu 1989; Nanbu et al. 2000; Simpson et al. 1995). These bacteria secret glucosyltransferases (GTFs) that convert dietary sucrose to high molecular weight glucan in a single step reaction: Sucrose + (glucan) n → (glucan) n + 1 + fructose. The glucans thus produced enhance the colonization of the bacteria and promote the formation of dental plaque on tooth surfaces (Hanada and Slade 1980). Unless otherwise stated, our use of the term “glucan” refers specifically to the bacterial-derived polymer.
There are at least three GTF encoding genes in the genome of S. mutans: gtfB, gtfC, and gtfD (Shiroza et al. 1987; Hanada and Kuramitsu 1989; Shiramura et al. 1994). The glucans produced by GTF-B and GTF-C are primarily water-insoluble (IG – Insoluble Glucans) and predominantly α-(1→3)-linked, while the GTF-D enzyme synthesizes water-soluble (SG – Soluble Glucans) glucan with mainly α-(1→6)-linkages. Mutagenesis studies performed on these genes revealed that converting the consensus amino acid residues in the IG-producing gtf genes to the corresponding residues of the SG-producing gtf gene resulted in mutant enzymes producing primarily SG and vice versa (Shimamura et al. 1994). The glucosyl linkages and primer dependency could also be altered through this mutagenesis strategy. These data demonstrate that the glucan polymers can be altered by mutating the GTF enzymes that make them. With various DNA evolution technologies available, one can imagine that glucans with diverse properties can be produced with designer enzymes to deliver specific functionality for various industrial and food applications.
Plants are efficient production systems for heterologous proteins that lead to high-value products (Kusnadi et al. 1997; Cairns 2003). Direct genetic modification of starch biosynthesis is complex and only limited success has been achieved because multiple genes are involved in the pathway. In contrast, production of glucan by GTFs can be carried out in a single-step enzymatic reaction from sucrose which is abundantly present in developing maize grains. Morever, unlike other polysaccharide-synthesizing enzymes, GTFs do not require nucleotide-activated sugars as biosynthetic intermediates. Taken together, the simplicity of the reaction, abundance of the sucrose substrate in plants, and the large demand for various glucan products make it economically attractive and practically feasible to produce this bacterial glucan in plants through genetic engineering. Several attempts have been made to modify potato tuber starch with transgenic approaches. By introducing an E. coli glycogen synthase into potato tuber amyloplast, highly branched starch was produced in the transgenic plant (Shewmaker et al. 1994). Unique carbohydrates were also synthesized in potato amyloplasts by expressing a cyclodextrin-glycosyltransferase (Oakes et al. 1991). Kok-Jacon et al. (2003) discussed an attempt to introduce new α-(1→3)- and α-(1→6) - linkages in potato starch with a glucansucrase/dextransucrase (also called GTF) transgenic approach. Recently, they successfully introduced the L. mesenteroides dextransucrase in potato; this resulted in a significant level of glucan accumulation in potato tuber juices (Kok-Jacon et al. 2005).
Like glucan, fructan consists of α-(1→6) linked fructose. Fructan is also synthesized from sucrose via a single enzymatic step catalyzed by fructosyltransferases whose reaction mechanisms are similar to GTFs (Chambert and Gonzy-Treboul 1976). Fructan biosynthesis was reported to be successfully induced in potato, sugar beet, maize, tobacco, and chicory by ectopic expression of fructosyltransferases from plant or bacterial sources (van der Meer et al. 1994; Caimi et al. 1996; Rober et al. 1996; Sevenier et al. 1998; Sprenger et al. 1997). Ebskamp et al. (1994) introduced the SacB gene from B. subtilis into tobacco, resulting in 3–8% fructan accumulation in the tobacco leaves; Caimi et al. (1996) transformed maize with the SacB gene from B. amyloliquefaciens, leading to fructan accumulation up to 4–8% of the dry weight of mature seeds.
In the present work, we developed transgenic maize endosperm that accumulates novel glucan polymers at levels relevant to commercial production. This was done through the introduction of a Streptococcus mutans gtfD gene into maize. Expression of that gene yielded fully functional GTF-D enzyme as shown by accumulation of novel soluble α-(1→6)-linked glucan at high levels in the mature maize kernels (up to 14% of their dry weight).
Materials and methods
DNA constructs and maize transformation
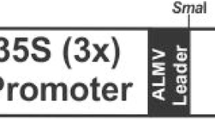
Standard procedures were used for DNA manipulation (Sambrook et al. 1989). The maize codon-optimized gtfD gene (GenBank accession number DQ659099) was cloned into a plant binary plasmid containing the ubiquitin promoter (Ub1, Christensen and Quail 1996) followed by the maize Brittle 1 transit peptide sequence (Bt1ts, Sullivan and Kaneko 1995) for amyloplast targeting, and potato protease inhibitor II 3′ noncoding sequence (pinII) as a terminator (Fig. 1, An et al. 1989). The resulting plasmid contained the BAR gene as a selectable marker and was used to transform freshly isolated immature Type II maize embryos (Armstong et al. 1991) through Agrobacterium-mediated transformation (Zhao et al. 2001). Basta-resistant calli were obtained after selection and maize plants were regenerated and grown to maturity in a greenhouse.
Schematic diagram of gtfD expression cassette in plasmid PHP11677 used for maize transformation. It contains the maize ubiquitin promoter (Ub1), maize ubiquitin intron 1, maize Brittle 1 transit peptide (TP), and potato protease inhibitor II terminator (pinII). The sizes of DNA fragments are indicated
Antibodies and Western blots
Polyclonal anti-GTF-D antibodies were raised in rabbits using SDS-PAGE purified proteins. After Commassie Brilliant Blue dye staining, the polyacrylamide gel slice containing the protein of interest was excised and homogenized with Adjuvant (Sigma, MO). The emulsified gel mixture was used to immunize the animals that had been prescreened for negative immuno-reactivity with maize proteins. Antibody thus raised had no detectable cross-reaction with other maize proteins. To detect GTF-D protein in maize, maize seedlings were homogenized in phosphate-buffered saline (PBS) buffer. After micro-centrifugation, total soluble protein extracts were fractionated in an 8% SDS polyacrylamide settle on this or SDS-PAGE and use one or the other consistently. The separated proteins were transferred to NC membrane. Western blotting was carried out with an ECL Kit (Amershane Pharmacia Biotech, NJ) according to the manufacture’s suggestions. Anti-GTF-D antibodies were diluted 1:2000 before use and goat anti-rabbit IgG (Sigma, MO) was used as the secondary antibody.
In vitro glucan synthesis
Crude E. coli cell extracts of approximate one milligram containing recombinant GTF-D wildtype or mutant proteins were added in a 2 ml reaction mixture containing 50 mM potassium phosphate, pH 6.5, 50 mM sucrose, and 0.02% sodium azide in a dialysis cassette (Pierce Chemical Company, IL). The reaction cassette was dialyzed against 500 ml of the same reaction mixture without the enzyme at 37°C overnight. Glucan synthesized inside the reaction cassette was precipitated with 75% methanol, washed three times with 75% methanol, and dried in air. The polymer was then further treated with proteinase K. The final glucan was re-precipitated, washed in 75% methanol and lyophilized.
Determination of molecular weight, water solubility, and the glucosyl-linkage of the in vitro synthesized glucans
The molecular weight of glucan polymer was determined by a low angle light scattering photometer (Wyatt Technology Corporation, CA) using polysaccharide molecular weight standards (JM Science, NY). Pure water was used as solvent and the measurement was carried out at room temperature. The relative water solubility was determined based on the amount of lyophilized glucan redissolved in distilled H2O. High solubility means the final glucan concentration is 10% or above; medium solubility refers to 1% to 10% of glucan in solution; and low solubility indicates 1% or less glucan in the redissolved solution. To determine the glucosyl-linkage, the in vitro synthesized glucans were first reduced for 7 d at 4°C with 4% (w/v) NaBH4 in 0.5 M NaOH, then neutralized with HCl, dialyzed against water, and lyophilized. The reduced glucans (30–60 mg) were dissolved in 0.5 M NaOH containing 10% (v/v) of D2O and 1% (w/v) sodium 4,4-dimethyl-4-silapentane-1-sulfonate, ultrasonicated, and analyzed by 13C nuclear magnetic resonance (NMR) spectrophotometry as described by Shimamura (1989).
Glucan detection and quantification in transgenic maize
A single dried mature corn kernel was powdered with a Tissue Raptor (Beckman, CA) at 1650 rpm. Twenty five milligram of the corn meal was digested with α-amylase (Sigma, MO) in 50 mM MOPS pH 7.0 at the ratio of 1:4 (mg corn meal: units of α-amylase). The reaction was carried out in a 2-ml screw-capped tube at 85°C overnight. Undigested polysaccharides were precipitated with 80% (v/v) ethanol. After washing the precipitates with 80% ethanol three times to remove monomeric and oligomeric sugars released from starch, the remaining meal was further digested with 30 units of dextranase (Worthington, NJ) in 100 mM KPi (pH 6.0) at 37°C for 2 h. The reaction was centrifuged and the supernatant was filtered through a Centricon-3 filter (Millipore, MA) to remove enzyme and undigested polymers. The filtrate thus obtained was analyzed for the presence of isomaltose (as described below). Isomaltose released from the meal after the second digestion was used to deduce the content of glucan. Glucan synthesized in vitro from the enzyme GTF-D/N471D was used as a standard in every analysis.
Isomaltose analysis
Analysis of isomaltose by HPLC was done on an anion exchange column PA1 (Dionex, CA) using a Dionex HPLC system DX500 equipped with an amperometric detector. Isomaltose eluted with 100 mM NaOH at 8.2 min at a flow rate of 1 ml/min. Its identification was judged with retention time using pure commercial available isomaltose (Sigma, MO) as a standard. In some cases, the identity of the putative isomaltose peak was confirmed by a mass spectrometer.
Starch granule size measurement
About 150 mg of ground corn meal was digested with pepsin (Sigma, MO) overnight at 37°C in 0.001 N HCl to release starch granules from the associated proteins (L. Zhou, personal communication). The digest was neutralized with 25 mM MOPS (pH 7.5) and then used to measure the median values of the granule size distribution with a Malvern 2000 Master Sizer (Malvern Instruments, MA).
Results
In vitro glucan synthesis and analysis
Glucans produced with different GTF enzymes from Streptococcus mutans have distinct physical properties. For example, GTF-B and GTF-C glucans are water-insoluble whereas GTF-D glucan is water-soluble. A single amino acid change in the catalytic domain of GTF-D enzyme was found to cause a dramatic alteration in the structures of the resulting glucans (Shimamura et al. 1994). To look for a suitable candidate gtf gene for plant transformation, we obtained the expression clones containing the wildtype and several mutant gtfD genes from Dr. Howard Kuramitsu in the State University of New York at Buffalo. The enzymes were expressed in E. coli and used to synthesize glucans from sucrose in vitro. Synthesis reactions were done in dialysis membranes to provide for the continuous supply of the substrate sucrose and concurrent removal of the reaction by-products, such as fructose, without diluting the enzyme. We found that the enzyme remained reactive for at least 72 h during the incubation and the yield of glucan synthesized was markedly increased with prolonged reaction. The synthesized glucans were analyzed for their glucose linkages, molecular weight, and water solubility as described in the Materials and Methods. The results for polymers derived from the wild-type GTF-D and two mutagenized enzymes, GTF-D/N471D and GTF-D/T589E, are summarized in Table 1. GTF-D/N471D glucan has the highest water solubility and molecular weight (MW 500,000), while GTF-D/T589E produced a smallest glucan molecule (MW 500) with the lowest water solubility. Wildtype GTF-D glucan was intermediate in both cases. All glucans contained mixed linkages of 1→2, 1→3, 1→4, 1→6, 1→3,6, and 1→4,6 though in quite differing ratios. The diversity of linkages provided a basis for the diverse properties of glucan polymers.
Based on the in vitro synthesis experiments, the quantity and MW of GTF-D/N471D-produced glucan was preferable to that of WT or GTF-D/T589E and, hence, it was chosen to be the transgenic enzyme for maize transformation.
Codon optimization and introduction of gtfD N471D into maize
In general, genes in monocots including maize contain a high G + C content at the degenerate third base; 16 out of 18 amino acids favor G or C in this position (Murray et al. 1989). However, bacterial genes, such as gtfD, follow a different codon bias pattern. It is advisable to optimize the bacterial gene’s codons to maize-preferred ones to achieve high levels of recombinant protein expression in transgenic plants. To wit, partially codon-optimized cryIA(b) gene resulted in 10-fold higher protein accumulation in plants relative to WT and fully codon-optimized cryIA(b) genes engendered 100-fold increases (Perlak et al. 1991). Increased recombinant protein accumulation due to codon optimization is reported in other cases (Horvath, et al. 2000 and Karasev et al. 2005). Following the maize codon usage guidelines of Murray et al. (1989), we optimized gtfD N471D with maize-preferred codons and produced a synthetic gene. The resulting sequence (Genbank accession number DQ659099), which has the exact same deduced amino acid sequence as the original enzyme, differs from the original bacterial gene in the following three ways: (1) the sequence encoding the 36 amino acid signal peptide was removed and replaced with ATG—an initiator methionine codon, (2) 29% of gtfD/N471D’s bases were changed according to maize preferred codon usage increase GC content from 38.5% to 62.8%, (3) several putative polyadenylation sites were eliminated. Additionally, we added the maize Brittle 1 (Bt1) transit peptide to the N-terminus of the enzyme, resulting in additional 234 nucleotides attached to the 5′-end of the maize-optimized gtfD/N471D sequence. The entire coding sequence was cloned into a plant binary plasmid behind the strong ubiquitin promoter (Fig. 1). Bt1 is an adenylate translocator on the amyloplast membrane whose transit peptide functions as a stromal targeting sequence (Shannon et al. 1998). In the present study, the gftD gene was constructed so that the enzyme would be targeted to amyloplasts, thus preventing any possible deleterious effects of glucan production on other cellular processes in the cytosol. Another advantage of compartmentalizing glucan synthesis in amyloplasts is that the foreign glucan could be co-accumulated with the naturally-occurring starch granules so that the well-established corn wet milling technology for starch extraction can be applied to glucan isolation.
The expression cassette was introduced into the Type II maize (Armstrong et al. 1991) via Agrobacterium tumefaciens-mediated transformation. The expression of gtfD gene was analyzed in the T0 seedlings by Western blot with the GTF-D specific antibodies. In all the events that expressed GTF-D, a 90 kDa fragment was detected; while one plant was identified to contain the full-length 120 kDa GTF-D, but it did not survive to maturity (Fig. 2). Both the 120 kDa and the 90 kDa bands contained the glucosyltransferase activity as was demonstrated with Periodic acid-Schiff base staining as described by Zugibe (1970). In this assay, the GTF-D enzyme in the polyacrylamide gel was buffered with 50 mM potassium phosphate (pH 6.5) containing 50 mM sucrose to synthesize glucan, which deposited where it was synthesized and reacted with periodic acid-Schiff base to form a white precipitate. We observed white precipitates in the same areas where the GTF-D Western positive bands were located (data not shown).
Western blot analysis of total proteins from PHP11677 transformed T0 seedling leaves. Protein samples from 8 independent transformation events (lanes 3–10), together with the untransformed wildtype control (lane 2), which is our elite maize line, were used for transformation. GTF-D specific antibodies were used. Lane 1 is protein molecular weight markers, whose sizes are indicated on the left (Invitrogen, CA). The 90 kD and 120 kD GTF-D bands are indicated with arrows
Glucan accumulation analysis in mature transgenic seeds
To identify and quantify glucans synthesized in maize, we developed an indirect assay to determine glucan accumulation in plant tissues where a high level of starch was present. Our assay utilized a dextranase (I.U.B.:3.2.1.11) to distinguish the structural differences between glucan and other maize endogenous polysaccharide polymers, especially starch. Dextranases catalyze the endohydrolysis of α-(1→6)-glucosidic linkages in dextran and produce isomaltose as the end product (Chaiet et al. 1970). The GTF-D glucan contains a preponderance of α-(1→6)-glucosidic linkages (Table 1) while starch is a polymer of α-[1→4] and α-[1→4,6] linkages. In the assay, α-amylase was used to digest the corn meal first to eliminate starch in the samples. This enzyme randomly hydrolyzes all the α-(1→4) glucosidic bonds in starch except the outermost ones and those next to branches, resulting in hydrolysis of starch to small molecules, which are not precipitated with ethanol. The remaining undigested GTF-D glucan was precipitated and further digested with dextranase to produce isomaltose, subsequently quantified by HPLC.
The retention time for the standard isomaltose (Sigma, MO) on an HPLC run was 8.2 min (data not shown). Transgenic corn meal digest had a peak that appeared at the same time (Fig. 3c). Such a peak did not exist for the non-transgenic parent, Type II corn meal (Fig. 3a). Adding the in vitro-synthesized glucan to the control Type II corn meal demonstrates the 8.2 min-peak again (Fig. 3b). To confirm the identity of this peak, we spiked the transgenic digest sample with isomaltose, which co-eluded with the 8.2 min peak (data not shown). Using the same detection procedure we analyzed non-transgenic maize kernels. No isomaltose after the second digestion could be detected, indicating that the isomaltose was indeed derived from glucan synthesized by the introduced GTF-D/N471D enzyme. Taken together, our results indicate that novel glucan has been produced in the gtfD N471D transgenic corn seeds.
After gtfD transformation, transgenic lines were self pollinated for five generations. We obtained 122 ears, all of which were homozygous for the gtfD transgene. We randomly picked one kernel from each ear and analyzed glucan content; results are shown in Fig. 4a. All transgenic seeds contained glucan ranging from 0.8% to 14% of the mature seeds’ dry weight. To determine glucan concentration variation, we analyzed the glucan content from 12 randomly—picked ears, 6 seeds from each ear. The results showed that there was glucan concentration variation within ears, although the variation was less pronounced than that between ears (Fig. 4b).
Detection of glucan accumulation in the gtfD transformed maize kernels. Glucan accumulation is expressed as weight percentage of whole kernel dry mass. (a) Each bar represents a randomly-picked kernel from 122 different transgenic ears. (b) Each bar represents the average glucan accumulation of 6 randomly-picked kernels from a single transgenic ear. Twelve ears were analyzed in total. Standard deviation among kernels is indicated as error bars
To explore the possibility that a higher level of available sucrose might increase the glucan accumulation in corn seeds, we generated maize lines whose progeny contain at least one copy of the gtfD gene and are homozygous for the sh2 recessive mutation. Sh2 encodes a subunit of endosperm ADP-glucose pyrophosphorylase, the key enzyme for starch biosynthesis in maize endosperm (Bhave et al. 1990). Mutations in Sh2 result in a great reduction in starch levels with concomitantly high sucrose accumulation. We hypothesized that the introduced GTF-D enzyme in the presence of elevated sucrose levels would cause commensurately higher glucan accumulation. In all the cases where the gtfD transgene was present in the sh2 background, glucan was detected. However, we failed to find increased glucan accumulation in the sh2 background (data not shown). One plausible explanation is that the elevated level of sucrose in the sh2 seeds might be compartmentalized in organelles other than amyloplasts where GTF-D was expected to be expressed. In other words, the excess sucrose under sh2 background might not be accessible to GTF-D enzyme. Further experiments to target GTF-D to various subcellular locations would be necessary in order to answer these questions.
Effects of glucan accumulation on plant morphology and starch granule size
The gtfD expressing plants that survived to maturity showed no phenotypic difference as compared with the untransformed control plants. The transgenic plants grew and developed normally during the life cycle both in greenhouse and in the field; they had no noticeable alteration in seed size, seed density, and opacity (data not shown). Since we targeted the production of glucan to amyloplasts where starch granules accumulate, we also determined the impact of glucan production on starch granule size. To this end, the starch granule size distribution was measured with a Malvern 2000 Master Sizer. There was no distinguishable difference in starch granule size distribution between the gtfD transformed and untransformed control starch (Fig. 5) and they had similar mean values in size (Table 2).
Discussion
In this report we demonstrated that introduction of bacterial glucosyltransferase gene gtfD into maize resulted in functional expression of the GTF-D protein and significant accumulation of glucan polymers in mature corn kernels. The glucan content in some transgenic kernels reached up to 14% of dry weight. Although there was a relatively low plant regenerating rate for the gtfD transgenic events (discussed below), the mature glucan-producing plants showed no noticeable phenotypic difference compared to wildtype plants, indicating the glucan polymers had no direct effect on plant and kernel development.
Quantifying glucan contents in transgenic plants is challenging. First, the enzymatic products are not homogeneous and exhibit a wide range in species-specific linkage and size distribution. Second, detection of glucan in maize seeds relies on effective methods of distinguishing between starch and glucan, which are difficult to separate physically. Third, the molecular weight of the glucans derived from GTF-D ranged from 30 kDa to 1,000 kDa. Its high molecular weight and heterogeneity made it impossible to resolve on a HPLC column which provides a more direct assay to measure polymers with a low degree of polymerization (Sevenier et al. 1998; this study). Kok-Jason et al. (2005) reported a method to detect dextrans in transgenic corn seeds with ELISA (enzyme-linked immunosorbent assay) using a monoclonal anti-α-(1→6) dextran antibody that recognizes the terminal non-reducing end of the polysaccharide. Our glucan quantification method took the advantage of the high percentage of α- (1→6) and the low percentage of α- (1→4) linkages present in the GTF-D glucan. The sequential enzymatic digestion (α-amylase followed by dextranase) of corn meal can effectively distinguish the endogenous corn starch in seeds from GTF-D glucan. The subsequent HPLC quantification of isomaltose, the end product of glucan dextranase digestion, is a very sensitive method. We obtained reproducible values for the same batch of samples, suggesting that our glucan quantification method is reliable for relative quantification. We note that our indirect quantification method depends on the essential equivalence of the glucan produced in vitro and in the endosperm.
To maximize the expression of the gtfD construct, we optimized the codon usage of gtfD with the maize optimal codons and used a strong promoter (Ub1) for gene expression. However, we were only able to detect a very low level of GTF-D protein in a 90 kDa truncated form instead of the expected 120 kDa full-length form. The GTF proteins have two main functional domains; the catalytic domain composed of about 900 amino acids in the N-terminus of the protein and a glucan-binding domain of about 500 amino acids in the C-terminus (Monchois et al. 1999). It is unlikely any part of the GTF-D catalytic domain could be truncated since it is essential for the enzyme activity. We know the 90 kDa truncated protein is functional based on its in vitro activity demonstrated with periodic acid-Schiff base staining and the presence of the glucan polymers in the transgenic plant. We suspect that the 90 kDa protein lost part of GTF-D’s C-terminus where the glucan-binding domain resides. No conclusive essential function has been assigned to that domain yet. At present, we do not know how the truncation happened. Further studies are needed to address this. That a low level of foreign enzyme expression still resulted in enzymatic product is not uncommon. It was reported that the SacB gene from Bacillus subtilis was expressed at a very low level: no corresponding mRNA and levansucrase protein could be detected in the transgenic tobacco plants even though a strong promoter and a translational enhancer were employed (Ebskamp et al. 1994). Yet the authors did detect fructan polymers, ranging from 3 to 8% of the dry weight.
Our gtfD transgene was driven by the strong ubiquitin promoter. However, we were only able to detect a very low level of gtfD mRNA by Northern Blot hybridization (data not shown). Since the ubiquitin promoter is also active in cultured cells, sucrose in the growth medium could be directed to polymer production in calli as they were being propagated and in the subsequent plant regeneration. We postulate that the presumbly high level of polymer production in calli is inimical to cellular robustness, leading to the recovery of lines only with relatively low GTF-D activity. This might also explain the low plant regenerating rate for gtfD transformation (3.2 %) versus a normal rate of 30% in general. Truncation of the GTF-D protein from 120 kDa to 90 kDa in our transgenic lines might be another way to keep the enzymatic activity low. It will be interesting to know if we can increase the expression of gtfD temporally and spatially by using different promoters, e.g. a seed specific promoter to activate GTF-D expression only during seed development to avoid any potential detrimental consequence of the enzyme expression during transgenic plant generation.
Maize lines deficient in ADPglucose pyrophosphorylase (AGPase)—for example, the sh2 mutant—accumulate sucrose to much higher levels than the cognate normals. As another attempt to maximize glucan production in transgenic maize plants, we out-crossed T1 transgenic plants with a sh2 maize line and followed by backcrossing. We obtained sh2 homozygous lines with at least one copy of the gtfD transgene hoping that the higher sucrose level in the sh2 background could be routed to glucan production. However, we failed to observe any obvious alteration in glucan accumulation in the gtfD sh2 background, suggesting that either the elevated level of sucrose was not routed to the amyloplast where GTF-D presumably functioned or sucrose level was not the limiting factor for a higher level of glucan production. Caimi et al. (1996) reported that a dramatic increase of fructan accumulation in the sh2 mutant when targeting a SacB gene in the maize endosperm vacuoles. There is no doubt that the vacuolar sucrose level is higher in the sh2 background as compared with that in the cognate wildtype lines. However, there is no published study concerning how cytosolic sucrose crosses membranes to get into amyloplasts. Whether the high level of cytosolic sucrose in the sh2 background could efficiently cross the amyloplast membrane without any help is still a question. The next phase of study will be introducing a sucrose transporter on the amyloplast membrane to pump in more sucrose for glucan biosynthesis by GTF-D enzyme. Such a mechanism may also be useful in a normal genetic background as well as sh2.
Free fructose formed during glucan biosynthesis may accumulate as an end product in the amyloplast and be unable to further participate in glycolysis or cytosolic sucrose synthesis. Fructose must be phosphorylated by either fructokinases or hexokinases before it can be further metabolized. Two maize fructokinase genes, ZmFRK1 and ZmFRK2, have been cloned in our lab (Zhang et al. 2003). Introducing them into the gtfD transgenic plant will possibly increase the kernel sink strength to pull higher amounts of sucrose during seed development. In addition, it might alleviate any possible adverse impacts that high fructose concentration might have on cells. All of these may contribute to enhance glucan production in corn seeds.
Here we have demonstrated a feasible approach to produce novel glucans in mature maize kernels from sucrose in a single-step, one-enzyme reaction. From the in vitro studies conducted in other labs as well as in our lab, we know that glucans with special functionality can be produced with proper modification of the GTF enzymes (Shimamura et al. 1994). A parallel study is ongoing to obtain modified GTF enzymes through gene-shuffling technology (Stemmer 1994) for novel glucan polymers production in plants (G. Zhu, personal communication). Due to maize’s large production capacity and the well-established wet milling technology, a large quantity of glucans with superior physical properties to the original bacterial glucans is within our reach.
References
An G, Mitra A, Choi HK, Costa MA, An K, Thornburg RW, Ryan CA (1989) Functional analysis of the 3′ control region of the potato wound-inducible proteinase inhibitor II gene. Plant Cell 1:115–122
Armstrong CL, Green CE, Phillips RL (1991) Development and availability of germplasm with a high Type II culture formation response. Maize Gen Coop Newslett 65:92–93
Bhave MR, Lawrence S, Barton C, Hannah LC (1990) Identification and molecular characterization of shrunken-2 cDNA clones of maize. Plant Cell 2:581–588
Blanshard JMV (1987) Starch granule structure and function: a physicochemical approach. In: Galliard T (ed) Starch: Properties and potential. John Wiley & Sons, New York, pp 17–53
Caimi PG, McCole LM, Klein TM, Kerr PS (1996) Fructan accumulation and sucrose metabolism in transgenic maize endosperm expressing a Bacillus amyloliquefacins SacB gene. Plant Physiol 110:55–363
Cairns AJ (2003) Fructan biosynthesis in transgenic plants. J Exp Bot 54:549–67
Chambert R, Conzy-Treboul G (1976) Levansucrase of Bacillus subtilis: characterization of a stabilized fructosyl-enzyme complex and identification of an aspartyl residue as the binding site of the fructosyl group. Eur J Biochem 71:493–508
Chaiet L, Kempf AJ, Harman R, Kaczka E, Weston R, Nollstadt K, Wolf FJ (1970) Isolation of a pure dextranase from Penicillium funiculosum. Appl Microbiol 20:421–6
Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5:213–218
Ebskamp MJM, van der Meer IM, Spronk BA, Weisbeek PJ, Smeekens CM (1994) Accumulation of fructose polymers in transgenic tobacco. Biotechnology 12:272–276
Hamada S, Slade HD (1980) Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev 44:331–384
Hanada N, Kuramitsu HK (1989) Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect Immun 57:2079–2085
Horvath H, Huang J, Wong O, Kohl E, Okita T, Kannangara CG, von Wettstein D (2000) The production of recombinant proteins in transgenic barley grains. Proc Natl Acad Sci USA 97:1914–1919
Karasev AV, Foulke S, Wellens C, Rich A, Shon KJ, Zwierzynski I, Hone D, Koprowski H, Reitz M (2005) Plant based HIV-1 vaccine candidate: Tat protein produced in spinach. Vaccine 23:1875–1880
Kok-Jacon GA, Ji Q, Vincken JP, Visser RG (2003) Towards a more versatile alpha-glucan biosynthesis in plants. J Plant Physiol 160:765–77
Kok-Jason GA, Vincken J-P, Suurs LCJM, Wang D, Liu S, Visser RGF (2005) Production of dextran in transgenic potato plants. Transgenic Res 14:385–395
Kusnadi AR, Nikolov ZL, Howard JA (1997) Production of recombinant proteins in transgenic plants: practical considerations. Biotechnol Bioeng 56:473–484
Monchois V, Willemot RM, Monsan P (1999) Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol Rev 23:131–151
Murray EE, Lotzer J, Eberle M (1989) Codon usage in plant genes. Nucleic Acids Res 17:477–498
Nanbu A, Hayakawa M, Takada K, Shinozaki N, Abiko Y, Fukushima K (2000) Production, characterization, and application of monoclonal antibodies which distinguish four glucosyltransferases from Streptococcus sobrinus. FEMS Immunol Med Microbiol 27:9–15
Oakes JV, Shewmaker CK, Stalker DM (1991) Production of cyclodextrins, a novel carbohydrate, in the tubers of transgenic potato plants. Biotechnology 9:982–986
Perlak FJ, Fuchs RL, Dean DA, McPherson SL, Fischhoff DA (1991) Modification of the coding sequence enchances plant expression of insect control protein genes. Proc Natl Acad Sci USA 88:3324–3328
Rober M, Geider K, Muller-Rober B, Willmitzer L (1996) Synthesis of fructans in tubers of transgenic starch-deficient potato plants does not result in an increased allocation of carbohydrates. Planta 199:528–536
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning – Laboratory manuals. Cold Spring Harbor Laboratory Press Painview, New York
Sevenier R, Hall RD, van der Meer IM, Hakkert HJ, van Tunen AJ, Koops AJ (1998) High level fructan accumulation in a transgenic sugar beet. Nat Biotechnol 16:843–846
Shannon JC, Pien FM, Cao H, Liu KC (1998) Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP-glucose into amyloplasts of maize endosperms. Plant Physiol 117:1235–1252
Shewmaker CK, Boyer CD, Wiesenborn DP, Thompson DB, Boersig MR, Oakes JV, Stalker DM (1994) Expression of Escherichia coli glycogen synthase in the tubers of transgenic potatoes (Solanum tuberosum) results in a highly branched starch. Plant Physiol 104:1159–1166
Shimamura A (1989) Use of 13C-NMR spectroscopy for the quantitative estimation of 3-O-. and 3,6-di-O-substituted d-glucopyranosyl residues in a-D-glucans formed by the D-glucosyltransferase of Streptococcus sobrinus. Carbohyd Res 185:239–248
Shimamura A, Nakano YJ, Mukasa H, Kuramitsu HK (1994) Identification of amino acid residues in Sreptococcus mutans glucosytransferases influencing the structure of the glucan product. J Bacteriol 176:4845–4850
Shiroza T, Ueda S, Kuramitsu HK (1987) Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol 169:4263–4270
Simpson CL, Cheetham NW, Giffard PM, Jacques NA (1995) Four glucosyltransferases, GtfJ, GtfK, GtfL and GtfM, from Streptococcus salivarius ATCC 25975. Microbiology, 141:1451–1460
Sprenger N, Schellenbaum L, van Dun K, Boller T, Wiemken A (1997) Fructan synthesis in transgenic tobacco and chicory plants expressing barley sucrose: fructan 6-fructosyltransferase. FEBS Lett 400:355–358
Stemmer WP (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389–391
Sullivan TD, Kaneko Y (1995) The maize brittle 1 gene encodes amyloplast membrane polypeptides. Planta 196:477–484
van der Meer IM, Ebskamp M, Visser R, Weisbeek PJ, Smeekems S (1994) Frucan as a new carbohydrate sink in transgenic potato plants. Plant Cell 6:561–570
Zhang S, Nichols SE, Dong JG (2003) Cloning and characterization of two fructokinases from maize. Plant Sci 165:1051–1058
Zhao ZY, Gu W, Cai T, Tagliani L, Hondred D, Bond D, Schroeder S, Rudert M, Pierce D (2001) High throughput genetic transformation mediated by Agrobacterium tumefaciens in maize. Mol Breed 8:323–333
Zugibe FT (1970) Positive periodic acid-Schiff staining of acid mucopolysaccharides. Histochem J 2:191–1977
Acknowledgements
We would like to thank Professor Howard Kuramitsu (State University of New York at Buffalo) for providing with the gtfD expression vectors, Teresa Harp and Michaela Owen for HPLC analysis of glucan dextranase digests, Lan Zhou for assistance with analysis of starch granule size distribution, and Xiaoming Bao and Keith Roesler for critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zhang, S., Dong, J.G., Wang, T. et al. High level accumulation of α-glucan in maize kernels by expressing the gtfD gene from Streptococcus mutans . Transgenic Res 16, 467–478 (2007). https://doi.org/10.1007/s11248-006-9049-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-006-9049-8