Abstract
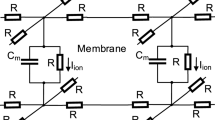
The cardiac bidomain model is a popular approach to study electrical behavior of tissues and simulate interactions between the cells by solving partial differential equations. The iterative and data parallel model is an ideal match for the parallel architecture of Graphic Processing Units (GPUs). In this study, we evaluate the effectiveness of architecture-specific optimizations and fine grained parallelization strategies, completely port the model to GPU, and evaluate the performance of single-GPU and multi-GPU implementations. Simulating one action potential duration (350 msec real time) for a 256×256×256 tissue takes 453 hours on a high-end general purpose processor, while it takes 664 seconds on a four-GPU based system including the communication and data transfer overhead. This drastic improvement (a factor of 2460×) will allow clinicians to extend the time-scale of simulations from milliseconds to seconds and minutes; and evaluate hypotheses in a shorter amount of time that was not feasible previously.
Similar content being viewed by others
References
Ten Tusscher KH, Panfilov AV (2006) Alternans and spiral breakup in a human ventricular tissue model. Am J Physiol 90:326–345
Jack JJB, Noble D, Tsien RW (1975) Electric current flow in excitable cells. Clarendon, Oxford
NVIDIA Corporation (2007), CUDA Programming Guide 1.0. http://www.nvidia.com
Micikevicius P (2009) 3d Finite difference computation on GPUs using CUDA. In: GPGPU-2: Proceedings of 2nd workshop on general purpose processing on graphics processing units. ACM, New York, pp 79–84
Qu Z, Garfinkel A (1999) An advanced numerical algorithm for solving partial differential equation in cardiac conduction. IEEE Trans Biomed Eng 46(9):1166–1168
Roth BJ (1991) Action potential propagation in a thick strand of cardiac muscle. Circ Res 68:162–173
Smith GD (1985) Numerical solution of partial differential equations: finite difference methods, 3rd edn. Clarendon, London
Ten Tusscher KHWJ, Noble D, Noble PJ, Panfilov AV (2004) A model for human ventricular tissue. Am J Physiol 286:H1573–H1589
Luo CH, Rudy Y (1991) A model of the ventricular cardiac action potential: depolarization, repolarization, and their interaction. Circ Res 68:1501–1526
Beeler GW, Reuter H (1977) Reconstruction of the action potential of ventricular myocardial fibres. J Physiol 268:177–210
Noble D (1962) A modification of the Hodgkin-Huxley equations applicable to Purkinje fibre action and pace-maker potentials. J Physiol 160:317–352
McAllister RE, Noble D, Tsien RW (1975) Reconstruction of the electrical activity of cardiac Purkinje fibres. J Physiol 251:1–59
Yanagihara K, Noma A, Irisawa H (1980) Reconstruction of sino-atrial node pacemaker potential based on the voltage clamp experiments. Jpn J Physiol 30:841–857
Cardiac modeling, http://www.cardiacmodeling.org/
Vigmond EJ, Boyle PM, Joshua Leon L, Plank Gernot (2009) Near-real-time simulations of biolelectric activity in small mammalian hearts using graphical processing units. In: Engineering in medicine and biology society, pp 3290–3293
Sato D, Xie Y, Weiss JN, Qu Z, Garfinkel A, Sanderson AR (2009) Acceleration of cardiac tissue simulation with graphic processing units. In: Medical and biological engineering and computing, pp 1011–1015
Unat D, Cai X, Baden S (2010) Optimizing the Aliev-Panfilov model of cardiac excitation on heterogeneous systems. University of Iceland, Reykjavik
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nimmagadda, V.K., Akoglu, A., Hariri, S. et al. Cardiac simulation on multi-GPU platform. J Supercomput 59, 1360–1378 (2012). https://doi.org/10.1007/s11227-010-0540-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-010-0540-x