Abstract
Backbone 15N relaxation parameters and 15N–1HN residual dipolar couplings (RDCs) have been measured for a variant of human α-lactalbumin (α-LA) in 4, 6, 8 and 10 M urea. In the α-LA variant, the eight cysteine residues in the protein have been replaced by alanines (all-Ala α-LA). This protein is a partially folded molten globule at pH 2 and has been shown previously to unfold in a stepwise non-cooperative manner on the addition of urea. 15N R2 values in some regions of all-Ala α-LA show significant exchange broadening which is reduced as the urea concentration is increased. Experimental RDC data are compared with RDCs predicted from a statistical coil model and with bulkiness, average area buried upon folding and hydrophobicity profiles in order to identify regions of non-random structure. Residues in the regions corresponding to the B, D and C-terminal 310 helices in native α-LA show R2 values and RDC data consistent with some non-random structural propensities even at high urea concentrations. Indeed, for residues 101–106 the residual structure persists in 10 M urea and the RDC data suggest that this might include the formation of a turn-like structure. The data presented here allow a detailed characterization of the non-cooperative unfolding of all-Ala α-LA at higher concentrations of denaturant and complement previous studies which focused on structural features of the molten globule which is populated at lower concentrations of denaturant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
While structural biology generally concerns itself with the study of folded states of proteins, the study of denatured, unfolded or partially-folded states is equally important. Understanding the structure and dynamics of these states can help to elucidate mechanisms and processes which occur during protein folding (Arai and Kuwajima 2000; Dinner et al. 2000; Ptitsyn 1995). Non-native states of proteins have also been recognized to be involved in a range of disease states (Dobson 2001; Kelly 2002). In addition, it has been shown that many proteins and protein domains are unstructured or only partially structured under physiological conditions (Wright and Dyson 1999). Non-native protein states are not characterized by a single stable three-dimensional structure. Instead they consist of ensembles of interconverting conformers. NMR is well suited to the study of such non-native protein states (Dyson and Wright 2001; Shortle 1996). Among the NMR techniques and parameters which have proved to be useful in defining conformational ensembles are chemical shift deviations (Eliezer et al. 2000; Ramboarina and Redfield 2003; Spera and Bax 1991; Wishart and Sykes 1994), 3JNHα coupling constants (Smith et al. 1996), short-range 1H–1H NOEs (Schwalbe et al. 1997), paramagnetic relaxation enhancement (Gillespie and Shortle 1997a, b), 15N relaxation measurements (Dyson and Wright 2001; Klein-Seetharaman et al. 2002; Schwalbe et al. 1997) and unfolding with urea (Schulman et al. 1997; Wang et al. 1995; Wang and Shortle 1995). Together with mutagenesis (Klein-Seetharaman et al. 2002; Nishimura et al. 2005; Song et al. 1998; Wu and Kim 1998) these have enabled the localization of short-range secondary structure propensities and long-range non-native hydrophobic clusters within unfolded and partially-folded proteins.
More recently, residual dipolar couplings (RDCs) have emerged as an important tool for characterizing residual structure in unfolded or partially-folded proteins (Meier et al. 2008). Initial studies by Shortle and co-workers on denatured staphylococcal nuclease, eglin c and short unstructured peptides showed these to have comparatively uniform non-zero RDC values (Ackerman and Shortle 2002a, b; Ohnishi et al. 2004; Ohnishi and Shortle 2003; Shortle and Ackerman 2001). Predictions using random flight and valence chains show this to be the expected behavior of a fully-denatured polypeptide chain (Louhivuori et al. 2004, 2003; Obolensky et al. 2007). Improved fits between experimental and predicted RDCs are obtained when using structural models derived from Φ/Ψ propensities of coil regions in folded proteins (Bernadó et al. 2005b; Jha et al. 2005). Deviations from these models can be used to identify regions of persistent residual structure. This approach has been used to characterize the populations of residual helical structure in ribonuclease S-peptide (Alexandrescu and Kammerer 2003), ACBP (Fieber et al. 2004), apomyoglobin (Mohana-Borges et al. 2004) and Sendai virus nucleoprotein (Jensen et al. 2008), to probe persistent long-range contacts in α-synuclein (Bernadó et al. 2005a) but lacking in β-synuclein (Bertoncini et al. 2007), to identify residues involved in persistent hydrophobic clusters or close to proline residues in a nucleocapsid-binding domain (Bernadó et al. 2005b), apomyoglobin (Mohana-Borges et al. 2004) and the A-state of foldon (Meier et al. 2004) and to locate turn conformations populated in denatured ubiquitin (Meier et al. 2007) and natively unfolded Tau protein (Mukrasch et al. 2007). In addition, variations in RDC values with temperature have been used to identify melting hotspots during the unfolding of the protein GB1 (Ding et al. 2004) and to characterize the unfolding of the β-hairpin in the A-state of foldon (Meier et al. 2004).
In this paper we use 15N relaxation and RDC measurements to study the stepwise unfolding in urea of a molten globule formed by a variant of human α-lactalbumin (α-LA) (Fig. 1). Non-native states of α-lactalbumin have been studied extensively using a wide range of experimental and theoretical approaches (Kuwajima 1996; Ramboarina and Redfield 2003, 2008; Redfield et al. 1999; Schulman et al. 1997, 1995; Smith et al. 2005; Song et al. 1998; Wu and Kim 1998). α-LA forms a molten globule, the so-called A-state, at low pH (Ptitsyn 1995). Heteronuclear NMR methods have shown that this A-state unfolds in a very non-cooperative manner on the addition of urea (Schulman et al. 1997). A core containing residues from the native-state A, B, D and C-terminal 310 helices in α-LA persists even in 10 M urea. α-LA contains four disulfide bonds. The unfolding of a variant of α-LA, in which the eight cysteine residues have been replaced by alanines (all-Ala α-LA), has been characterized using the same approach (Redfield et al. 1999). Interestingly, this variant shows a similar persistent core and non-cooperative unfolding on the addition of urea. However, in all-Ala α-LA the core containing residues from the A, B, C, D and C-terminal 310 helices only persists up to concentrations of 3 M urea. Unfolding is virtually complete in 7 M urea, as indicated by the observation of all expected peaks in the 15N–1H HSQC spectrum; circular dichroism measurements at 222 nm show that almost all helical structure is lost in 7 M urea (Redfield et al. 1999). In this paper, we investigate the all-Ala α-LA variant by measuring 15N relaxation and RDC data for the protein at pH 2 in 4, 6, 8 and 10 M urea. By combining these data with 15N chemical shifts, results from urea unfolding experiments and studies of small peptides we have been able to derive a comprehensive picture of the backbone dynamics and residual structure present during protein unfolding.
Schematic representation of the native structure of human α-lactalbumin. The α- and β-domains and the five α-domain helices are labelled. The diagram was generated using MOLSCRIPT (Kraulis 1991)
Materials and methods
15N-labelled all-Ala α-LA was expressed and purified as described elsewhere for related proteins (Peng et al. 1995; Schulman et al. 1995). Samples for the 15N relaxation and RDC measurements were prepared at a protein concentration of 0.4 mM in 4, 6, 8 and 10 M urea at pH 2. 6% polyacrylamide gels were used to align the samples for RDC measurements. These were prepared in a similar manner to that described by Sass et al. (2000). The isotropic data were collected first using Shigemi microcells. The dried gels were then added to the samples, and allowed to swell before being compressed from an initial length of 3 cm to a final length of 2.1 cm. In this manner it was possible to ensure that the urea concentration remained the same for both datasets. Repeat experiments were conducted in order to estimate the error. Average RDC values are reported and the error was found to be ~0.5 Hz at all urea concentrations. Despite this proportionately fairly large error, the magnitude and sign of RDCs was reproduced extremely well in separate datasets, even when using different samples.
NMR experiments were performed on a home-built NMR spectrometer, located in the Department of Biochemistry, operating at a 1H frequency of 599.48 MHz. 15N–1HN RDCs were measured at 20°C using an HSQC experiment which incorporated an S3E pulse-sequence element (Meissner et al. 1997). 128 and 1,024 complex points and sweep widths of 1,272.26 and 6,756.76 Hz were collected in F1 (15N) and F2 (1H), respectively. The data were processed using Felix 2.3 (Accelrys) and zero-filled to give final digital resolutions of 0.3 Hz/pt and 3.3 Hz/pt in F1 and F2, respectively. 15N R1 and R2 relaxation rates and the {1H}–15N NOE were measured using standard pulse sequences at 20°C, with 128 and 1,024 complex points and sweep widths of 1,152.07 and 6,514.66 Hz in F1 (15N) and F2 (1H), respectively. The R1 and R2 measurements used a series of 9 to 12 experiments with relaxation delays ranging from 20 to 2,500 ms and from 8.6 to 1,200 ms, respectively. The {1H}–15N NOE is reported as the ratio of peak heights in spectra collected with and without saturation of the 1H spectrum by a non-selective 90° pulse every 10 ms for a period of 4 s. Errors in the measurement of peak heights were estimated from baseplane noise. Fitting of R1 and R2 rates and reduced spectral density mapping (Bracken et al. 1999; Farrow et al. 1995) were carried out using in-house programs written in Fortran77.
An ensemble of 5,000 all-Ala α-LA structures was generated using the Jha et al. algorithm (http://unfolded.chicago.edu/) (Jha et al. 2005). RDCs were predicted for this ensemble using the program PALES (Zweckstetter and Bax 2000). Each structure was aligned separately and the resulting RDCs were then averaged over the ensemble and scaled so that the mean predicted RDC value was the same as that observed experimentally. The average area buried upon folding (AABUF) (Rose et al. 1985) and hydrophobicity (Abraham and Leo 1987) profiles for all-Ala α-LA were calculated using the ExPASy web server (http://expasy.org) weighted over a 7-residue window. The bulkiness profile for all-Ala α-LA was calculated as described by Cho et al. (2007).
Results
15N relaxation data for all-Ala α-LA
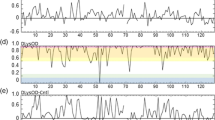
R1 and R2 relaxation rates and the {1H}–15N heteronuclear NOE ratio measured in 4, 6, 8 and 10 M urea are plotted as a function of the all-Ala α-LA sequence in Fig. 2 (the full data are provided as Electronic supplementary material). The relaxation data at high urea concentrations are similar to those reported for other denatured proteins (Schwalbe et al. 1997; Schwarzinger et al. 2002). The R2 relaxation rates, generally the most informative of the three parameters for denatured proteins, are smaller at the termini and rise to a plateau in the central region of the polypeptide chain. This is in agreement with predictions from a simple model of an unfolded polypeptide chain with unrestrained segmental motion in which the 15N relaxation properties of a given amide group are not influenced to any significant extent by the identity of neighboring residues in the chain (Schwalbe et al. 1997). Deviations from this pattern are largest at 4 and 6 M urea and decrease as the urea concentration is increased. In 10 M urea, average R2 values of 4.89 and 4.81 are observed for the α-domain (residues 1–39 and 82–123) and the β-domain (residues 40–81), respectively. Elevated R2 relaxation rates are observed in the α-domain of the protein, particularly in 4 M urea. A sharp decrease in R2 is observed around residue 20, particularly in 6, 8, and 10 M urea. The sequence from 17–20 is GYGG and the high conformational freedom of glycine residues presumably enables this region to be relatively more flexible at all urea concentrations. In addition, residues 17–22 correspond to a loop region between the native A and B helices (Fig. 1). This loop region has been observed to unfold at a lower urea concentration than the flanking helices (Redfield et al. 1999). A less pronounced decrease in R2 is observed in the vicinity of Gly35 at the end of the B helix. The differing plateau values for R2 at the three highest urea concentrations reflect a viscosity effect; as the urea concentration is raised, the viscosity of the sample increases, the overall molecular tumbling is slowed and the R2 relaxation rate is raised. Interestingly, the data collected at 4 M urea do not follow this trend and the R2 values are higher than expected. Furthermore, residues 40–60 form a higher plateau than residues 61–80 which could be the result of differential exchange broadening in the β-domain in 4 M urea. This would correlate with previous NMR studies of all-Ala α-LA which have shown that the protein is not completely unfolded in 4 M urea (Redfield et al. 1999) and a recent study of backbone dynamics of the all-Ala α-LA molten globule which has shown differences in fast timescale dynamics within the β-domain; residues 40–60 show more restricted mobility than residues 61–80 (Ramboarina and Redfield 2008).
15N relaxation data for all-Ala α-LA plotted as a function of the amino acid sequence. a {1H}–15N NOE as Isat/Inonsat, b R1 and c R2 values for all-Ala α-LA in 4 M (black), 6 M (red), 8 M (cyan) and 10 M (gold) urea are shown. All data were collected at 600 MHz and 20°C. The secondary structure and domain organization found in native α-lactalbumin is summarized above
15N relaxation data for folded proteins are usually analyzed using the model-free formalism of Lipari and Szabo; using this approach order parameters and internal correlation times can be extracted for individual residues (Lipari and Szabo 1982a, b). This approach is not appropriate for unfolded proteins because such systems cannot be described by an overall rotational correlation time. Instead, reduced spectral density mapping is often used to describe backbone motions in unfolded proteins (Farrow et al. 1997; Le Duff et al. 2006; Schwarzinger et al. 2002; Shojania and O’Neil 2006; Wirmer et al. 2006b) and this approach has been adopted here for all-Ala α-LA (Fig. 3). Both J(0.87ωH) and J(ωN) show little variation with amino acid sequence and urea concentration. Some deviations in these spectral densities are observed for α-domain residues in 4 M urea but these are associated with large errors due to the very weak peak intensities. J(0), which is most sensitive to R2, shows a significant dependence on the urea concentration for residues located in the α-domain of the protein. The increased J(0) values in the α-domain observed as the urea concentration is lowered reflect the increasing contributions from exchange processes on the millisecond to microsecond time scale as the protein becomes more structured.
15N chemical shift data for all-Ala α-LA
15N chemical shifts measured in 6, 8 and 10 M urea are in good agreement with the sequence-corrected shifts predicted by Braun et al. (1994); correlation coefficients of 0.98 are obtained for comparison of predicted and experimental 15N shifts in both 6 and 10 M urea. Nevertheless, differences in 15N chemical shifts are observed as a function of urea concentration in the spectra of all-Ala α-LA (Redfield et al. 1999). The difference between the 15N chemical shifts measured in 10 and 6 M urea is plotted as a function of sequence in Fig. 4a. Chemical shift differences as large as 1 ppm are observed. The largest differences are observed in the α-domain in regions of sequence that correspond to the native A, B, D and C-terminal 310 helices. These are the regions of the protein that showed the largest exchange contributions to R2 at lower urea concentrations. The chemical shifts for these regions in 6 M urea are shifted upfield of the 10 M values, consistent with the presence of residual helical propensity at the lower urea concentration (Wishart and Sykes 1994). For residues 17–23, which are located between the native A and B helices, much smaller chemical shift differences are observed; this is consistent with the unfolding of this loop at a lower concentration of urea. Smaller chemical shift differences are observed in the β-domain but the pattern is not uniform; somewhat larger differences are observed for residues 40–60 than observed for 61–80. The larger shift differences observed for residues 53, 54 and 59 are consistent with the recent observation of restricted backbone dynamics for residues in this region of the all-Ala α-LA molten globule (Ramboarina and Redfield 2008). A large negative shift difference is observed for Val66; this residue precedes a proline in the sequence.
a Difference in 15N chemical shifts for all-Ala α-LA at 6 and 10 M urea at 20°C [δ15N(10 M)–δ15N(6 M)]. b–e Experimental RDCs recorded for all-Ala α-LA in compressed polyacrylamide gels at 20°C (black) are compared with predicted RDCs for all-Ala α-LA based on a coil-model (red). Measurements were made in b 4 M, c 6 M, d 8 M and e 10 M urea. The secondary structure and domain organization found in native α-lactalbumin is summarized above
Experimental RDCs for denatured all-Ala α-LA
15N–1HN RDCs were measured for all-Ala α-LA in compressed polyacrylamide gels in the presence of 4, 6, 8 and 10 M urea at pH 2 and 20°C. The chemical shifts observed in the compressed gels are identical to those in isotropic solution, suggesting that the structural ensemble of all-Ala α-LA is not perturbed in the polyacrylamide gel medium. The RDC values are plotted as a function of amino acid sequence in Fig. 4b–e (Tables listing the RDC values are available as Electronic supplementary material). The RDC values are positive for all four datasets with average values of 0.55, 0.95, 1.17 and 1.11 Hz in 4, 6, 8 and 10 M urea, respectively. The observation of positive non-zero RDC values for denatured all-Ala α-LA in compressed polyacrylamide gels is in agreement with predictions for random flight and valence chains (Louhivuori et al. 2004, 2003) and data reported for other denatured proteins (Fieber et al. 2004; Meier et al. 2004; Mohana-Borges et al. 2004). Deviations from uniform values across the sequence may be due to nearest neighbor effects (Jha et al. 2005) and the inherent Φ/Ψ preferences of this amino acid sequence (Bernadó et al. 2005b; Louhivuori et al. 2004). Alternatively it may be indicative of specific long-range interactions favoured by the polypeptide chain (Bernadó et al. 2005a). This will be explored in the next section using a coil model for all-Ala α-LA. The absence of sustained stretches of negative RDC values indicates that no persistent helical structure is present (Alexandrescu and Kammerer 2003; Fieber et al. 2004), which is consistent with circular dichroism data for all-Ala α-LA in urea solutions (Redfield et al. 1999).
Although the pattern of RDC values for the protein at different urea concentrations is similar overall, there are some significant differences. For instance in 6 M urea negative values are observed for Asp16 and Gly17. In 8 M urea the value for Asp16 is positive and in 10 M urea both residues have positive values. By contrast, negative RDC values are observed for Tyr 103, Trp 104 and Leu 105 in 6, 8 and 10 M urea. In 4 M urea, RDC values for the majority of α-domain residues could not be measured because the peaks are too broad. The correlation coefficients between the larger data sets are 0.70 (6 and 8 M urea), 0.64 (8 and 10 M urea) and 0.46 (6 and 10 M urea); this is considerably lower than found for staphylococcal nuclease in water and at different urea concentrations (Shortle and Ackerman 2001) or for ACBP at 2.5 and 4 M GuHCl (Fieber et al. 2004). This suggests that although the protein is largely unfolded at the higher urea concentrations used, significant changes take place in the structural ensemble of all-Ala α-LA as the urea concentration is increased from 4 to 10 M; this is consistent with the previous observation of stepwise non-cooperative unfolding of all-Ala α-LA in urea (Redfield et al. 1999).
Modelling RDCs for denatured all-Ala α-LA
Two all-atom models have been used to predict RDC values for completely unfolded proteins (Bernadó et al. 2005b; Jha et al. 2005). Here we have used an ensemble of 5,000 structures generated using the Jha et al. coil algorithm including nearest-neighbor effects (Jha et al. 2005) and have calculated average predicted RDCs for this ensemble as described in the “Materials and methods” section. Deviations from these predicted values should indicate regions of residual structure within all-Ala α-LA. Figure 4b–e show comparisons of the experimental RDCs with those predicted from the statistical coil model. The correlation coefficients between experimental and calculated RDCs are 0.06, 0.17 and 0.25 at 6, 8 and 10 M urea, respectively. It is likely that residues with a negative experimental RDC value are involved in residual structure; after removing these values the correlation coefficients increase to 0.16, 0.32 and 0.50. The improvement of the correlation between the experimental and calculated values at higher urea concentration reflects the increasingly unfolded conformational ensemble for all-Ala α-LA. The correlation coefficient of 0.50 obtained for the 10 M urea RDC data is significantly lower than that reported by Jha et al. for apomyoglobin (0.71) and ubiquitin (0.70) but higher than the values for eglin c (0.30) and Δ131Δ staphylococcal nuclease (0.14) (Jha et al. 2005).
Differences between the experimental data and the predicted RDCs from the coil model can be broadly placed into two categories. The first of these includes residues 25–35 (the B helix in native α-LA) and the C-terminal residues (310-helix in native α-LA): the calculated RDC values deviate significantly from the experimental values measured in 6 M and to some extent in 8 M urea, but match those from 10 M urea well. This is accompanied by R2 values in these parts of the sequence which are elevated in 4 M urea but decrease to the plateau value in 10 M urea. These residues are evidently involved in residual structure at lower urea concentration, but become unstructured at high urea concentrations. A second group is formed by residues 101–106 (D helix in native α-LA) which do not correlate well with the predicted RDC values at any urea concentration, and show somewhat elevated R2 values even in 10 M urea. This indicates that these residues are to some extent structured, even at this high urea concentration.
Other discrepancies between the experimental and predicted RDCs can be attributed to certain amino acid types and sequence characteristics. Higher than expected RDCs are observed around the two proline residues (Pro24 and Pro67), for example, which suggests that the coil model for all-Ala α-LA does not sufficiently account for the way in which proline rigidifies the polypeptide chain. Wells et al. made a similar observation that their initial statistical coil model did not adequately model the conformational behaviour around proline residues (Wells et al. 2008). The low RDC values for the highly mobile GYGG sequence between residues 17 and 20, on the other hand, are well predicted by the model. Interestingly, the tryptophan residues (Trp60, Trp104 and Trp118) give rise to regions of lower RDC values than predicted and in many cases to negative values. While sustained stretches of RDCs with the opposite sign have been shown to be characteristic of helical conformations, it has been shown that changes in sign spanning just a couple of residues can arise from turn conformations (Mukrasch et al. 2007). Model peptides containing residues 101–111 and 111–120 from α-LA were shown to adopt turn-like structures for residues 103–106 and residues 115–118, respectively (Demarest et al. 1999; Demarest and Raleigh 2000). Thus, Trp104 and Trp118 may be inducing turns in the all-Ala α-LA structure even in the presence of denaturant. To investigate the possibility of long-range contacts, different ensemble sub-sets of the random-coil ensemble were considered (Bernadó et al. 2005a), but no evidence of long-range contacts was found for all-Ala α-LA which is consistent with previous studies (Wirmer et al. 2006a).
Discussion
Previous studies have elucidated the pattern of non-cooperative unfolding of the α-LA molten globule as the concentration of urea is increased (Schulman et al. 1997). Chemical shifts were used to monitor this unfolding event by exploiting the observation that in the molten globule state the NMR resonances are very broad and cannot in general be resolved. This is due to fluctuations on a millisecond to microsecond time scale in the populated conformational ensemble. On unfolding, however, sharp well-resolved resonances appear in the NMR spectrum. The urea-induced unfolding is, therefore, followed by monitoring the appearance of these sharp resonances in the spectra as the concentration of urea is increased. The results of these studies for all-Ala α-LA show that the residues that are most resistant to urea-induced unfolding are those in the regions corresponding to helices A, B, C, D and the C-terminal 310 helix of native α-LA (Redfield et al. 1999). The HSQC cross peaks of many of the residues in these regions are only observed when the protein is exposed to urea concentrations of 4 M or higher. The observed pattern of the appearance of peaks in HSQC spectra indicates that unfolding of the all-Ala α-LA molten globule in urea is a non-cooperative process.
Here we have extended the investigation of all-Ala α-LA by measuring 15N relaxation data and RDC values at urea concentrations of 4, 6, 8 and 10 M. It is now possible to characterize the unfolding of all-Ala α-LA in greater detail and, in particular, to follow the unfolding of residual structure occurring at higher urea concentrations. Both the 15N relaxation and RDC data show that the region corresponding to the native D helix unfolds last and is involved in non-random interactions even in 10 M urea. The native A and B helices are the next most stable parts of the structure, with millisecond time scale motions present in 8 M urea as well as RDCs indicative of non-random coil conformations. The regions of all-Ala α-LA corresponding to the C and C-terminal 310 helices of native α-LA show non-random interactions up to a urea concentration of 6 M.
Other evidence for persistent non-random structure in the C-terminal region of the protein comes from studies of short peptide fragments (residues 101–120) taken from the α-LA sequence (Demarest et al. 1998, 1999, 2001; Demarest and Raleigh 2000). These have identified two hydrophobic clusters involving residues 101–104 and 115–119, respectively. The cluster involving Ile101, Tyr103 and Trp104 has been shown to be intact in 8 M urea and based on the 15N relaxation and RDC data we propose that this remains intact even in 10 M urea in all-Ala α-LA. The negative RDC values in this region are consistent with the formation of a turn-like structure by these residues (Mukrasch et al. 2007). It has been shown that ubiquitin forms a hairpin turn at urea concentrations as high as 8 M (Meier et al. 2007).
It has been a long-term goal to be able to predict non-random structure directly from the polypeptide sequence. It has been shown previously that amino-acid bulkiness averaged over a 5-residue window along the protein sequence is able to predict RDCs for unfolded polypeptides (Cho et al. 2007). The experimental RDC data for all-Ala α-LA in 10 M urea are compared with predictions from the bulkiness profile for the all-Ala α-LA sequence in Fig. 5a. A reasonable correlation is obtained for most regions of the sequence with the exception of residues 103–106. The trends in the bulkiness profile along the sequence are similar to those in the predicted RDC values from the statistical coil model although the bulkiness profile is smoother as the values are averaged over a five-residue window. The good agreement between the measured RDCs and the bulkiness profile is not surprising when high concentrations of denaturant are present, since the effect of electrostatic and hydrophobic forces within the polypeptide chain will be minimized by the denaturant. The only physicochemical property of the amino acids which remains a dominant factor is their relative size (or bulk).
a Experimental RDCs recorded for all-Ala α-LA in 10 M urea (black) are compared with the bulkiness profile (red) calculated as described by Cho et al. (2007). b Normalised values for all Ala α-LA of the average area buried upon unfolding (AABUF) (black) (Rose et al. 1985) are compared with the hydrophobicity using the Abraham & Leo scale (red) (Abraham and Leo 1987). All values were averages over 7-residue windows. The dashed grey line shows the mean value of AABUF and hydrophobicity
As the concentration of denaturant is lowered, electrostatic and hydrophobic forces will start to come into play. In studies of apomyoglobin, the average area buried upon folding (AABUF) has been suggested to provide a good indication of those regions within the polypeptide chain which might be involved in the initial non-random interactions which then initiate the collapse and folding of the protein (Nishimura et al. 2005; Rose et al. 1985). An alternative parameter which has been investigated in this context is hydrophobicity (Wirmer et al. 2006a). Hydrophobicity scales generally disregard amino acid size; lysine and arginine, for instance, are assigned low values on the basis that they are charged. Significantly higher values are attributed to amino acids such as alanine, which although small is entirely hydrophobic (Abraham and Leo 1987). AABUF, on the other hand, takes size into account and assigns lower values to Ala than Arg or Lys (Rose et al. 1985).
The AABUF and hydrophobicity profiles for all-Ala α-LA are compared in Fig. 5b; these two parameters show similar trends in most regions of the sequence. The regions corresponding to the native B, D and C-terminal 310 helices, which unfold at the highest urea concentrations, are predicted well by both parameters. The R2 data and 15N chemical shift differences between 6 and 10 M urea in the region of residues 15–25 indicate significant backbone mobility and a lack of residual structure, particularly for residues 17–20; this is better reflected by the AABUF than the hydrophobicity values.
Surprisingly, comparison of the experimental data and the AABUF and hydrophobicity values for the A-helix region and the first half of the β-domain shows less agreement. The experimental data suggest that the A-helix region is involved in residual structure at urea concentrations of 6–8 M urea, but the AABUF and hydrophobicity values in this region are only slightly larger than the average values across the whole sequence. It may be the high degree of helical propensity rather than the hydrophobicity in this region of the sequence which is driving the formation of residual structure. The experimental R2 values and 15N chemical shifts suggest that the β-domain is largely unfolded at urea concentrations of 6 M and higher. Reasonably good agreement is observed between the experimental RDCs and both the values predicted from the statistical coil model (Fig. 4b–e) and the bulkiness profile (Fig. 5a) confirming that the β-domain is largely unfolded in 6 M urea. The AABUF and hydrophobicity profiles on the other hand show elevated values within the β-domain, in particular for residues 50–60. The higher values for residues ~50–60 compared to residues ~61–70 mirror the slightly elevated R2 values which are observed for residues 40–60 compared to residues 61–80 in 4 M urea and correlate with a recent study of backbone dynamics of the all-Ala α-LA molten globule that has shown more restricted mobility for residues 40–60 than for 61–80 (Ramboarina and Redfield 2008). This suggests that some non-random behavior may be present in the β-domain at lower urea concentrations but that this structure does not persist at higher urea concentrations as it does in the α-domain. The correlation between measured NMR parameters and the AABUF profile although reasonable for all-Ala α-LA, does not appear to be as clear as that reported previously for apomyoglobin (Nishimura et al. 2005).
In conclusion, the measurement of 15N relaxation data and RDCs for all-Ala α-LA at several concentrations of urea has provided a more detailed understanding of the stepwise non-cooperative unfolding of this protein at higher urea concentrations. This has identified the presence of non-random interactions at urea concentrations as high as 10 M which arise from conformational sampling of turn structures. The presence of such transient structure was not evident from the previous HSQC-based urea titration study which focused largely on the loss of stable secondary structure and long-range hydrophobic interactions in the molten globule which gave rise to significant peak broadening at lower urea concentrations (Redfield et al. 1999). The application of both approaches can provide a more complete picture of the loss of structure that occurs as a molten globule unfolds in urea.
References
Abraham DJ, Leo AJ (1987) Extension of the fragment method to calculate amino acid zwitterion and side chain partition coefficients. Proteins 2:130–152
Ackerman MS, Shortle D (2002a) Molecular alignment of denatured states of staphylococcal nuclease with strained polyacrylamide gels and surfactant liquid crystalline phases. Biochemistry 41:3089–3095
Ackerman MS, Shortle D (2002b) Robustness of the long-range structure in denatured staphylococcal nuclease to changes in amino acid sequence. Biochemistry 41:13791–13797
Alexandrescu AT, Kammerer RA (2003) Structure and disorder in the ribonuclease S-peptide probed by NMR residual dipolar couplings. Protein Sci 12:2132–2140
Arai M, Kuwajima K (2000) Role of the molten globule state in protein folding. Adv Prot Chem 53:209–282
Bernadó P, Bertoncini CW, Griesinger C, Zweckstetter M, Blackledge M (2005a) Defining long-range order and local disorder in native α-synuclein using residual dipolar couplings. J Am Chem Soc 127:17968–17969
Bernadó P, Blanchard L, Timmins P, Marion D, Ruigrok RW, Blackledge M (2005b) A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc Natl Acad Sci USA 102:17002–17007
Bertoncini CW, Rasia RM, Lamberto GR, Binolfi A, Zweckstetter M, Griesinger C, Fernandez CO (2007) Structural characterization of the intrinsically unfolded protein β-synuclein, a natural negative regulator of α-synuclein aggregation. J Mol Biol 372:708–722
Bracken C, Carr PA, Cavanagh J, Palmer AGIII (1999) Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol 285:2133–2146
Braun D, Wider G, Wüthrich K (1994) Sequence-corrected 15N random coil chemical shifts. J Am Chem Soc 116:8466–8469
Cho MK, Kim HY, Bernado P, Fernandez CO, Blackledge M, Zweckstetter M (2007) Amino acid bulkiness defines the local conformations and dynamics of natively unfolded α-synuclein and tau. J Am Chem Soc 129:3032–3033
Demarest SJ, Raleigh DP (2000) Solution structure of a peptide model of a region important for the folding of α-lactalbumin provides evidence for the formation of nonnative structure in the denatured state. Proteins 38:189–196
Demarest SJ, Fairman R, Raleigh DP (1998) Peptide models of local and long-range interactions in the molten globule state of human α-lactalbumin. J Mol Biol 283:279–291
Demarest SJ, Hua Y, Raleigh DP (1999) Local interactions drive the formation of nonnative structure in the denatured state of human α-lactalbumin: a high resolution structural characterization of a peptide model in aqueous solution. Biochemistry 38:7380–7387
Demarest SJ, Horng J-C, Raleigh DP (2001) A protein dissection study demonstrates that two specific hydrophobic clusters play a key role in stabilizing the core structure of the molten globule state of human α-lactalbumin. Proteins 42:237–242
Ding K, Louis JM, Gronenborn AM (2004) Insights into conformation and dynamics of protein GB1 during folding and unfolding by NMR. J Mol Biol 335:1299–1307
Dinner AR, Sali A, Smith LJ, Dobson CM, Karplus M (2000) Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem Sci 25:331–339
Dobson CM (2001) The structural basis of protein folding and its links with human disease. Phil Trans R Soc Lond, B 356:133–145
Dyson HJ, Wright PE (2001) Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol 339:258–270
Eliezer D, Chung J, Dyson HJ, Wright PE (2000) Native and non-native secondary structure and dynamics in the pH 4 intermediate of apomyoglobin. Biochemistry 39:2894–2901
Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE (1995) Spectral density function mapping using 15N relaxation data exclusively. J Biomol NMR 6:153–162
Farrow NA, Zhang O, Forman-Kay JD, Kay LE (1997) Characterization of the backbone dynamics of folded and denatured states of an SH3 domain. Biochemistry 36:2390–2402
Fieber W, Kristjansdottir S, Poulson FM (2004) Short-range, long-range and transition state interactions in the denatured state of ACBP from residual dipolar couplings. J Mol Biol 339:1191–1199
Gillespie JR, Shortle D (1997a) Characterization of long-range structure in the denatured state of staphylococcal nuclease. I. Paramagnetic relaxation enhancement by nitroxide spin labels. J Mol Biol 268:158–169
Gillespie JR, Shortle D (1997b) Characterization of long-range structure in the denatured state of staphylococcal nuclease. II. Distance restraints from paramagnetic relaxation and calculation of an ensemble of structures. J Mol Biol 268:170–184
Jensen MR, Houben K, Lescop E, Blanchard L, Ruigrok RW, Blackledge M (2008) Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: application to the molecular recognition element of Sendai virus nucleoprotein. J Am Chem Soc 130:8055–8061
Jha AK, Colubri A, Freed KF, Sosnick TR (2005) Statistical coil model of the unfolded state: resolving the reconciliation problem. Proc Natl Acad Sci USA 102:13099–13104
Kelly JW (2002) Towards an understanding of amyloidogenesis. Nat Struct Biol 9:323–325
Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H (2002) Long-range interactions within a nonnative protein. Science 295:1719–1722
Kraulis PK (1991) MOLSCRIPT: a program to produce detailed and schematic plots of protein structures. J Appl Crystallog 24:946–950
Kuwajima K (1996) The molten globule state of α-lactalbumin. FASEB J 10:102–109
Le Duff CS, Whittaker SB-M, Radford SE, Moore GR (2006) Characterisation of the conformational properties of urea-unfolded Im7: implications for the early stages of protein folding. J Mol Biol 364:824–835
Lipari G, Szabo A (1982a) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc 104:4546–4559
Lipari G, Szabo A (1982b) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc 104:4559–4570
Louhivuori M, Pääkkönen K, Fredriksson K, Permi P, Lounila J, Annila A (2003) On the origin of residual dipolar couplings from denatured proteins. J Am Chem Soc 125:15647–15650
Louhivuori M, Fredriksson K, Pääkkönen K, Permi P, Annila A (2004) Alignment of chain-like molecules. J Biomol NMR 29:517–524
Meier S, Güthe S, Kiefhaber T, Grzesiek S (2004) Foldon, the natural trimerization domain of T4 fibritin, dissociates into a monomeric A-state form containing a stable β-hairpin: atomic details of trimer dissociation and local β-hairpin stability from residual dipolar couplings. J Mol Biol 344:1051–1069
Meier S, Strohmeier M, Blackledge M, Grzesiek S (2007) Direct observation of dipolar couplings and hydrogen bonds across a beta-hairpin in 8 M urea. J Am Chem Soc 129:754–755
Meier S, Blackledge M, Grzesiek S (2008) Conformational distributions of unfolded polypeptides from novel NMR techniques. J Chem Phys 128:052204
Meissner A, Duus JØ, Sørensen OW (1997) Spin-state-selective excitation. Application for E.COSY-type measurement of JHH coupling constants. J Magn Reson 128:92–97
Mohana-Borges R, Goto NK, Kroon GJA, Dyson HJ, Wright PE (2004) Structural characterisation of unfolded states of apomyoglobin using residual dipolar couplings. J Mol Biol 340:1131–1142
Mukrasch MD, Markwick P, Biernat J, von Bergen M, Bernadó P, Griesinger C, Mandelkow E, Zweckstetter M, Blackledge M (2007) Highly populated turn conformations in natively unfolded tau protein identified from residual dipolar couplings and molecular simulation. J Am Chem Soc 129:5235–5243
Nishimura C, Lietzow MA, Dyson HJ, Wright PE (2005) Sequence determinants of a protein folding pathway. J Mol Biol 351:383–392
Obolensky OI, Schlepckow K, Schwalbe H, Solov’yov AV (2007) Theoretical framework for NMR residual dipolar couplings in unfolded proteins. J Biomol NMR 39:1–16
Ohnishi S, Shortle D (2003) Observation of residual dipolar couplings in short peptides. Proteins 50:546–551
Ohnishi S, Lee AL, Edgell MH, Shortle D (2004) Direct demonstration of structural similarity between native and denatured Eglin c. Biochemistry 43:4064–4070
Peng Z, Wu LC, Kim PS (1995) Local structural preferences in the α-lactalbumin molten globule. Biochemistry 34:3248–3252
Ptitsyn OB (1995) Molten globule and protein folding. Adv Prot Chem 47:83–229
Ramboarina S, Redfield C (2003) Structural characterisation of the human α-lactalbumin molten globule at high temperature. J Mol Biol 330:1177–1188
Ramboarina S, Redfield C (2008) Probing the effect of temperature on the backbone dynamics of the human α-lactalbumin molten globule. J Am Chem Soc 130:15318–15326
Redfield C, Schulman BA, Milhollen MA, Kim PS, Dobson CM (1999) α-Lactalbumin forms a compact molten globule in the absence of disulfide bonds. Nat Struct Biol 6:948–952
Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH (1985) Hydrophobicity of amino acid residues in globular proteins. Science 229:834–838
Sass H-J, Musco G, Stahl SJ, Wingfield PT, Grzesiek S (2000) Solution NMR of proteins within polyacrylamide gels: diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J Biomol NMR 18:303–309
Schulman BA, Redfield C, Peng Z, Dobson CM, Kim PS (1995) Different subdomains are most protected from hydrogen exchange in the molten globule and native states of human α-lactalbumin. J Mol Biol 253:651–657
Schulman BA, Kim PS, Dobson CM, Redfield C (1997) A residue-specific NMR view of the non-cooperative unfolding of a molten globule. Nat Struct Biol 4:630–634
Schwalbe H, Fiebig KM, Buck M, Jones JA, Grimshaw SB, Spencer A, Glaser SJ, Smith LJ, Dobson CM (1997) Structural and dynamical properties of a denatured protein. Heteronuclear 3D NMR experiments and theoretical simulations of lysozyme in 8 M Urea. Biochemistry 36:8977–8991
Schwarzinger S, Wright PE, Dyson HJ (2002) Molecular hinges in protein folding: the urea-denatured state of apomyoglobin. Biochemistry 41:12681–12686
Shojania S, O’Neil JD (2006) HIV-1 Tat is a natively unfolded protein. J Biol Chem 281:8347–8356
Shortle D (1996) Structural analysis of non-native states of proteins by NMR methods. Curr Opin Struct Biol 6:24–30
Shortle D, Ackerman MS (2001) Persistence of native-like topology in a denatured protein in 8 M urea. Science 293:487–489
Smith LJ, Bolin KA, Schwalbe H, MacArthur MW, Thornton JM, Dobson CM (1996) Analysis of main chain torsion angles in proteins: prediction of NMR coupling constants for native and random coil conformations. J Mol Biol 255:494–506
Smith LJ, Jones RM, van Gunsteren WF (2005) Characterization of the denaturation of human α-lactalbumin in urea by molecular dynamics simulations. Proteins 58:439–449
Song J, Bai P, Luo L, Peng Z (1998) Contribution of individual residues to formation of the native-like tertiary topology in the α-lactalbumin molten globule. J Mol Biol 280:167–174
Spera S, Bax A (1991) Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc 113:5490–5492
Wang Y, Shortle D (1995) The equilibrium folding pathway of staphylococcal nuclease: identification of the most stable chain–chain interactions by NMR and CD spectroscopy. Biochemistry 34:15895–15905
Wang Y, Alexandrescu AT, Shortle D (1995) Initial studies of the equilibirum folding pathway of staphylococcal nuclease. Phil Trans R Soc Lond, B 348:27–34
Wells M, Tidow H, Rutherford TJ, Markwick P, Ringkjøbing Jensen M, Mylonas E, Svergun DI, Blackledge M, Fersht AR (2008) Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci USA 105:5762–5767
Wirmer J, Berk H, Ugolini R, Redfield C, Schwalbe H (2006a) Characterization of the unfolded state of bovine α-lactalbumin and comparison with unfolded states of homologous proteins. Protein Sci 15:1397–1407
Wirmer J, Peti W, Schwalbe H (2006b) Motional properties of unfolded ubiquitin: a model for a random coil protein. J Biomol NMR 35:175–186
Wishart DS, Sykes BD (1994) Chemical shifts as a tool for structure determination. Methods Enzymol 239:363–392
Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293:321–331
Wu LC, Kim PS (1998) A specific hydrophobic core in the α-lactalbumin molten globule. J Mol Biol 280:1750182
Zweckstetter M, Bax A (2000) Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J Am Chem Soc 122:3791–3792
Acknowledgments
We thank Abishek Jha for providing an ensemble of 5,000 coil structures of all-Ala α-LA. C.R. is funded by the Wellcome Trust (Grant number 079440). V.A.H. was funded by a BBSRC graduate studentship and a bursary from St. Peter’s College.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
10858_2009_9342_MOESM1_ESM.pdf
Tables of heteronuclear NOE ratios, relaxation rates, spectral density values and RDCs for all-Ala α-LA in 4, 6, 8 and 10 M urea
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Higman, V.A., Rösner, H.I., Ugolini, R. et al. Probing the urea dependence of residual structure in denatured human α-lactalbumin. J Biomol NMR 45, 121–131 (2009). https://doi.org/10.1007/s10858-009-9342-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9342-y