Abstract
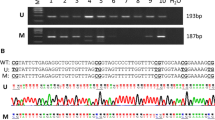
Breast Cancer Metastasis Suppressor 1 (BRMS1) suppresses metastasis of human breast cancer, ovarian cancer and melanoma in athymic mice. Studies have also shown that BRMS1 is significantly downregulated in some breast tumors, especially in metastatic disease. However, the mechanisms which regulate BRMS1 expression are currently unknown. Upon examination of the BRMS1 promoter region by methylation specific PCR (MSP) analysis, we discovered a CpG island (−3477 to −2214), which was found to be hypermethylated across breast cancer cell lines. A panel of 20 patient samples analyzed showed that 45% of the primary tumors and 60% of the matched lymph node metastases, displayed hypermethylation of BRMS1 promoter. Furthermore, we found a direct correlation between the methylation status of the BRMS1 promoter in the DNA isolated from tissues, with the loss of BRMS1 expression assessed by immunohistochemistry. There are several studies investigating the mechanism by which BRMS1 suppresses metastasis; however thus far there is no study that reports the cause(s) of loss of BRMS1 expression in aggressive breast cancer. Here we report for the first time that BRMS1 is a novel target of epigenetic silencing; and aberrant methylation in the BRMS1 promoter may serve as a cause of loss of its expression.




Similar content being viewed by others
Abbreviations
- 5-aza-dC:
-
5-Aza-2′-deoxycytidine
- BGS:
-
Bisulfite genomic sequencing
- BRMS1:
-
Breast Cancer Metastasis Suppressor
- IDV:
-
Integrated density value
- LN Met:
-
Metastatic breast cancer cells that metastasized to the lymph node
- MSP:
-
Methylation specific PCR
- TE:
-
Tris–EDTA
References
Shevde LA, Welch DR (2003) Metastasis suppressor pathways—an evolving paradigm. Cancer Lett 198:1–20. doi:10.1016/S0304-3835(03)00304-5
Samant RS, Seraj MJ, Saunders MM, Sakamaki TS, Shevde LA, Harms JF et al (2000) Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis 18:683–693. doi:10.1023/A:1013124725690
Seraj MJ, Samant RS, Verderame MF, Welch DR (2000) Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res 60:2764–2769
Samant RS, Debies MT, Shevde LA, Verderame MF, Welch DR (2002) Identification and characterization of the murine ortholog (brms1) of breast-cancer metastasis suppressor 1 (BRMS1). Int J Cancer 97:15–20. doi:10.1002/ijc.1569
Zhang Z, Yamashita H, Toyama T, Yamamoto Y, Kawasoe T, Iwase H (2006) Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin Cancer Res 12:6410–6414. doi:10.1158/1078-0432.CCR-06-1347
Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J (2005) Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases. J Cancer Res Clin Oncol 131:191–198. doi:10.1007/s00432-004-0629-9
Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP et al (2006) Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease-free survival in subsets of breast cancer patients. Clin Cancer Res 12:6702–6708. doi:10.1158/1078-0432.CCR-06-0635
Saunders MM, Seraj MJ, Li Z, Zhou Z, Winter CR, Welch DR et al (2001) Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res 61:1765–1767
Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ et al (2002) Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res 273:229–239. doi:10.1006/excr.2001.5452
DeWald DB, Torabinejad J, Samant RS, Johnston D, Erin N, Shope JC et al (2005) Metastasis suppression by breast cancer metastasis suppressor 1 involves reduction of phosphoinositide signaling in MDA-MB-435 breast carcinoma cells. Cancer Res 65:713–717
Liu Y, Smith PW, Jones DR (2006) Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol Cell Biol 26:8683–8696. doi:10.1128/MCB.00940-06
Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G (2005) Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-kappaB activity. Cancer Res 65:3586–3595. doi:10.1158/0008-5472.CAN-04-3139
Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH et al (2007) Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer 6:6. doi:10.1186/1476-4598-6-6
Hurst DR, Mehta A, Moore BP, Phadke PA, Meehan WJ, Accavitti MA et al (2006) Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone. Biochem Biophys Res Commun 348:1429–1435. doi:10.1016/j.bbrc.2006.08.005
Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL et al (2004) Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem 279:1562–1569. doi:10.1074/jbc.M307969200
Champine PJ, Michaelson J, Weimer BC, Welch DR, Dewald DB (2007) Microarray analysis reveals potential mechanisms of BRMS1-mediated metastasis suppression. Clin Exp Metastasis 24:551–565. doi:10.1007/s10585-007-9092-8
Cicek M, Samant RS, Kinter M, Welch DR, Casey G (2004) Identification of metastasis-associated proteins through protein analysis of metastatic MDA-MB-435 and metastasis-suppressed BRMS1 transfected-MDA-MB-435 cells. Clin Exp Metastasis 21:149–157. doi:10.1023/B:CLIN.0000024729.19084.f0
Rivera J, Megias D, Bravo J (2007) Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res 6:4006–4018
Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN et al (2001) Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat 65:101–110. doi:10.1023/A:1006461422273
Miller FR, Santner SJ, Tait L, Dawson PJ (2000) MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst 92:1185–1186. doi:10.1093/jnci/92.14.1185A
Ellison G, Klinowska T, Westwood RF, Docter E, French T, Fox JC (2002) Further evidence to support the melanocytic origin of MDA-MB-435. Mol Pathol 55:294–299. doi:10.1136/mp. 55.5.294
Rae JM, Ramus SJ, Waltham M, Armes JE, Campbell IG, Clarke R et al (2004) Common origins of MDA-MB-435 cells from various sources with those shown to have melanoma properties. Clin Exp Metastasis 21:543–552. doi:10.1007/s10585-004-3759-1
Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD (2007) MDA-MB-435 cells are derived from M14 melanoma cells–a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat 104:13–19. doi:10.1007/s10549-006-9392-8
Christgen M, Lehmann U (2007) MDA-MB-435: the questionable use of a melanoma cell line as a model for human breast cancer is ongoing. Cancer Biol Ther 6:1355–1357
Sellappan S, Grijalva R, Zhou X, Yang W, Eli MB, Mills GB et al (2004) Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res 64:3479–3485. doi:10.1158/0008-5472.CAN-3299-2
Welch DR (1997) Technical considerations for studying cancer metastasis in vivo. Clin Exp Metastasis 15:272–306. doi:10.1023/A:1018477516367
Hurst DR, Xie Y, Vaidya KS, Mehta A, Moore BP, Accavitti-Loper MA, Samant RS, Saxena R, Silveira AC, Welch DR (2008) Alterations of breast cancer metastasis suppressor 1:At rich interactive domain 4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem 283(12):7438–7444
Takai D, Jones PA (2003) The CpG island searcher: a new WWW resource. In Silico Biol 3:235–240
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics (Oxford, England) 18:1427–1431. doi:10.1093/bioinformatics/18.11.1427
Shevde LA, Samant RS, Paik JC, Metge BJ, Chambers AF, Casey G et al (2006) Osteopontin knockdown suppresses tumorigenicity of human metastatic breast carcinoma, MDA-MB-435. Clin Exp Metastasis 23:123–133. doi:10.1007/s10585-006-9013-2
Hartsough MT, Clare SE, Mair M, Elkahloun AG, Sgroi D, Osborne CK et al (2001) Elevation of breast carcinoma Nm23-H1 metastasis suppressor gene expression and reduced motility by DNA methylation inhibition. Cancer Res 61:2320–2327
Loeb DM, Evron E, Patel CB, Sharma PM, Niranjan B, Buluwela L et al (2001) Wilms’ tumor suppressor gene (WT1) is expressed in primary breast tumors despite tumor-specific promoter methylation. Cancer Res 61:921–925
Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B et al (2001) Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res 61:2782–2787
Umbricht CB, Evron E, Gabrielson E, Ferguson A, Marks J, Sukumar S (2001) Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene 20:3348–3353. doi:10.1038/sj.onc.1204438
Asch BB, Barcellos-Hoff MH (2001) Epigenetics and breast cancer. J Mammary Gland Biol Neoplasia 6:151–152. doi:10.1023/A:1011306222533
Yang X, Yan L, Davidson NE (2001) DNA methylation in breast cancer. Endocr Relat Cancer 8:115–127. doi:10.1677/erc.0.0080115
Jacinto FV, Esteller M (2007) Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis 22:247–253. doi:10.1093/mutage/gem009
Jing F, Zhang J, Tao J, Zhou Y, Jun L, Tang X et al (2007) Hypermethylation of tumor suppressor genes BRCA1, p16 and 14-3-3sigma in serum of sporadic breast cancer patients. Onkologie 30:14–19. doi:10.1159/000096892
Cheng L, Pan CX, Zhang JT, Zhang S, Kinch MS, Li L et al (2004) Loss of 14-3-3sigma in prostate cancer and its precursors. Clin Cancer Res 10:3064–3068. doi:10.1158/1078-0432.CCR-03-0652
Henrique R, Jeronimo C, Hoque MO, Carvalho AL, Oliveira J, Teixeira MR et al (2005) Frequent 14-3-3 sigma promoter methylation in benign and malignant prostate lesions. DNA Cell Biol 24:264–269. doi:10.1089/dna.2005.24.264
Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H et al (2000) High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA 97:6049–6054. doi:10.1073/pnas.100566997
Iwata N, Yamamoto H, Sasaki S, Itoh F, Suzuki H, Kikuchi T et al (2000) Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene 19:5298–5302. doi:10.1038/sj.onc.1203898
Osada H, Tatematsu Y, Yatabe Y, Nakagawa T, Konishi H, Harano T et al (2002) Frequent and histological type-specific inactivation of 14-3-3sigma in human lung cancers. Oncogene 21:2418–2424. doi:10.1038/sj.onc.1205303
Kaneuchi M, Sasaki M, Tanaka Y, Shiina H, Verma M, Ebina Y et al (2004) Expression and methylation status of 14-3-3 sigma gene can characterize the different histological features of ovarian cancer. Biochem Biophys Res Commun 316:1156–1162. doi:10.1016/j.bbrc.2004.02.171
Lombardi G, Di Cristofano C, Capodanno A, Iorio MC, Aretini P, Isola P et al (2007) High level of messenger RNA for BRMS1 in primary breast carcinomas is associated with poor prognosis. Int J Cancer 120:1169–1178. doi:10.1002/ijc.22379
Kelly LM, Buggy Y, Hill A, O′Donovan N, Duggan C, McDermott EW et al (2005) Expression of the breast cancer metastasis suppressor gene, BRMS1, in human breast carcinoma: lack of correlation with metastasis to axillary lymph nodes. Tumour Biol 26:213–216. doi:10.1159/000086955
Schwientek T, Nomoto M, Levery SB, Merkx G, van Kessel AG, Bennett EP et al (1999) Control of O-glycan branch formation. Molecular cloning of human cDNA encoding a novel beta1, 6-N-acetylglucosaminyltransferase forming core 2 and core 4. J Biol Chem 274:4504–4512. doi:10.1074/jbc.274.8.4504
Campbell IG, Phillips WA, Choong DY (2006) Genetic and epigenetic analysis of the putative tumor suppressor km23 in primary ovarian, breast, and colorectal cancers. Clin Cancer Res 12:3713–3715. doi:10.1158/1078-0432.CCR-06-0800
Gumy-Pause F, Wacker P, Maillet P, Betts DR, Sappino AP (2006) ATM promoter analysis in childhood lymphoid malignancies: a brief communication. Leuk Res 30:335–337. doi:10.1016/j.leukres.2005.07.012
Alvarez S, Diaz-Uriarte R, Osorio A, Barroso A, Melchor L, Paz MF et al (2005) A predictor based on the somatic genomic changes of the BRCA1/BRCA2 breast cancer tumors identifies the non-BRCA1/BRCA2 tumors with BRCA1 promoter hypermethylation. Clin Cancer Res 11:1146–1153
Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT (2002) Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene 21:4231–4236. doi:10.1038/sj.onc.1205528
Lu R, Fang JY, Zhu HY, Chen YX, Cheng ZH, Li EL (2004) Effect of eukaryotic expression plasmid DNA methyltransferase gene on methylation status and transcription level of DNA mismatch repair genes in human colon cancer cell line. Zhonghua Yi Xue Za Zhi 84:1014–1017
Wang L, Wang J, Sun S, Rodriguez M, Yue P, Jang SJ et al (2006) A novel DNMT3B subfamily, DeltaDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Int J Oncol 29:201–207
Tate PH, Bird AP (1993) Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev 3:226–231. doi:10.1016/0959-437X(93)90027-M
Sato M, Horio Y, Sekido Y, Minna JD, Shimokata K, Hasegawa Y (2002) The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene 21:4822–4829. doi:10.1038/sj.onc.1205581
Gao Y, Guan M, Su B, Liu W, Xu M, Lu Y (2004) Hypermethylation of the RASSF1A gene in gliomas. Clin Chim Acta 349:173–179. doi:10.1016/j.cccn.2004.07.006
Fang JY, Cheng ZH, Chen YX, Lu R, Yang L, Zhu HY et al (2004) Expression of Dnmt1, demethylase, MeCP2 and methylation of tumor-related genes in human gastric cancer. World J Gastroenterol 10:3394–3398
Steeg PS, Ouatas T, Halverson D, Palmieri D, Salerno M (2003) Metastasis suppressor genes: basic biology and potential clinical use. Clin Breast Cancer 4:51–62
Acknowledgments
This work was supported by grants BCTR0402317 (LAS) and BCTR0503488 (RSS) from Susan G. Komen for the Cure and CA88728 (DRW) and CA89019 (DRW, ARF, LRS, RSS), National Foundation for Cancer Research (DRW, ARF).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Metge, B.J., Frost, A.R., King, J.A. et al. Epigenetic silencing contributes to the loss of BRMS1 expression in breast cancer. Clin Exp Metastasis 25, 753–763 (2008). https://doi.org/10.1007/s10585-008-9187-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-008-9187-x